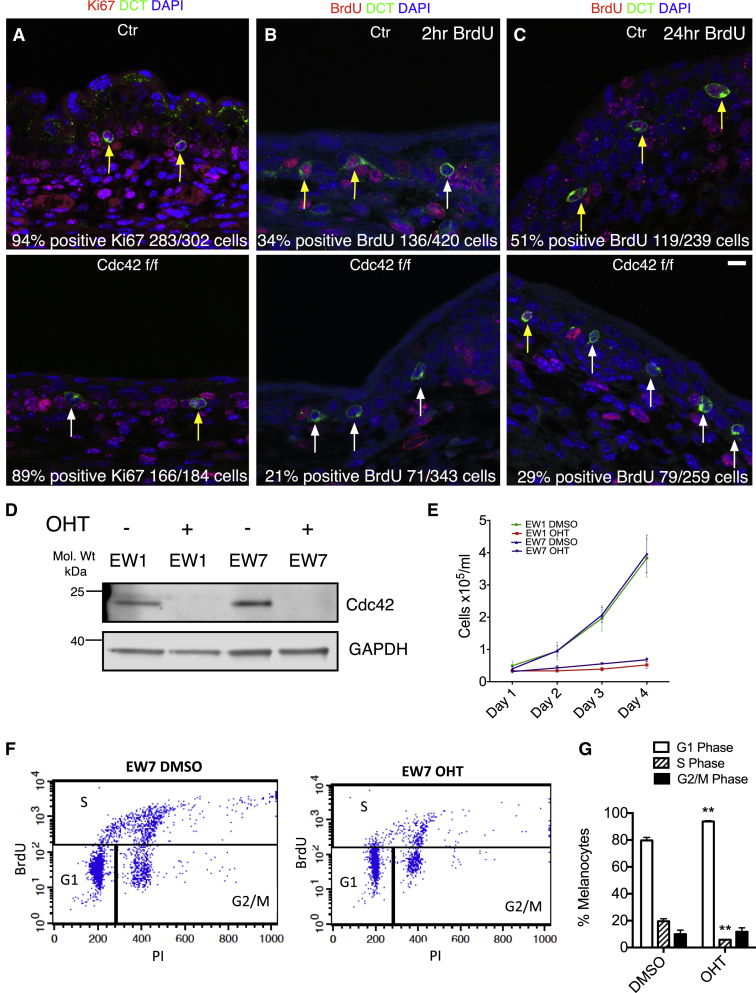
Figure 3.
Cdc42 Null Melanoblasts and Melanocytes Show Cell-Cycle Progression Defects
(A–C) Representative images of transverse sections of control (Ctr, Cdc42 wt/wt; Tyr::CreB; Z/EG; top) and Cdc42 f/f (Cdc42 f/f; TyrCreB; Z/EG; bottom) E15.5 embryo skin stained with DAPI (blue), DCT (green), and either Ki67 (red) or BrdU (red) at 2 or 24 hr as indicated. Yellow arrows indicate proliferating melanoblasts; white arrows indicate non-proliferating melanoblasts. N ≥ 3 separate embryos. t test with Welch’s correction. Scale bar, 10 μm.
(A) Ki67-positive cells as indicated; n.s., p = 0.38.
(B) 2 hr BrdU-positive cells as indicated; n.s., p = 0.06.
(C) 24 hr BrdU-positive cells as indicated; ∗p = 0.014.
(D) Western blot showing OHT-induced Cdc42 deletion (GAPDH loading control) in EW1 and EW7 Cdc42 f/f; ROSA26::Cre-ERT2; CDKN2−/−cells.
(E) Melanocyte proliferation assay of control (DMSO-treated) and Cdc42 f/f (OHT-treated) cell lines (EW1 and EW7). Error bars show SEM from N = 3 experiments in triplicate.
(F) Representative plot from flow-cytometry analysis of EW7 following mock treatment (DMSO) or deletion (OHT) of Cdc42.
(G) Quantification of cells in cycle. N = 3. Error bars show SEM. ∗∗p = 0.0039 (G1 phase), ∗∗p = 0.0017 (S phase).
See also Figure S3.