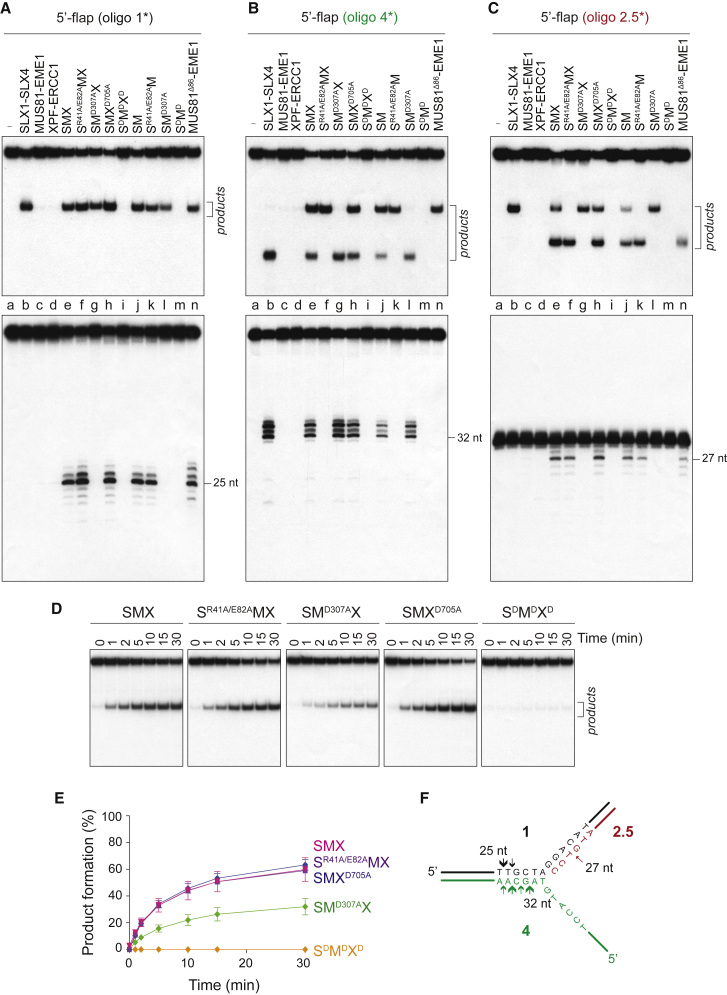
Figure 5.
In SMX, MUS81-EME1 Is Activated to Cleave 5′-Flap Substrates
(A–C) The 5′-flap substrate (10 nM), 5′-32P end-labeled in oligonucleotide 1 (A), 4 (B), or 2.5 (C) was incubated with the indicated enzyme (0.5 nM) for 5 min at 37°C. The reaction was divided in half and analyzed by native (top) and denaturing (bottom) PAGE. Incision sites were determined by comparison to 5′-32P end-labeled oligonucleotides of identical sequence and defined lengths. Asterisks denote the oligonucleotide that was 5′-32P end-labeled.
(D) Time course analysis of 5′-flap (50 nM) cleavage by wild-type and catalytically impaired SMX complexes (0.5 nM) containing mutations in SLX1 (SR41A/E82AMX), MUS81 (SMD307AX), XPF (SMXD705A), or all three nucleases (SDMDXD). The 5′-flap DNA was 5′-32P end-labeled in oligonucleotide 1 (F). Reaction products were analyzed by native PAGE.
(E) Quantification of (D). Cleavage products are expressed as a percentage of total radiolabeled DNA and represent the mean of at least three independent experiments. Error bars are SEM.
(F) Schematic of the 5′-flap DNA showing the main positions of incision by SMX. Arrow size represents relative incision efficiency (i.e., larger arrows indicate more efficient cut sites).
See also Figures S5 and S6.