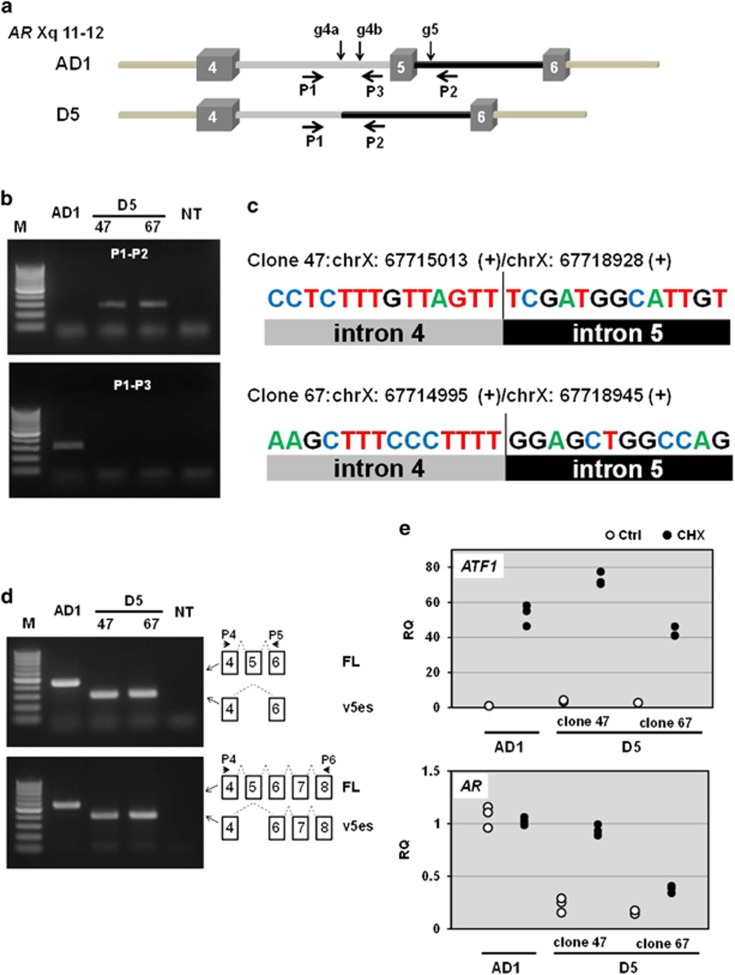
Figure 6.
ARv5es transcript may elicit NMD. (a) AR genomic structures featuring exons 4, 5 and 6 and the corresponding introns are drawn. Activation of non-homologous end joining pathway by CRISPER/Cas9 system results in deletion of exon 5 in AR gene in the parental AD1 cell to create cell line D5. While co-targeting with gRNAs g4a and g5 resulted in production of D5 clone 47, g4b and g5 were used to obtain clone 67. The information about gRNA is provided in Table 1. (b) PCR analysis of exon 5 deletion of AR gene. Genomic DNA isolated from AD1 and D5 clone 47 and 67 were used for PCR with primers P1 and P2 to test the absence of exon 5, and P1 and P3 primers were used as described in Table 1 to examine the absence of the targeted segment of intron 4 in D5 clones and its presence in AD1. M, DNA size marker (O'RangeRuler 100 bp DNA Ladder: Thermo Fisher Scientific). NT, non-template. (c) Direct Sanger DNA sequencing of the deletion-specific PCR products determined the breakpoints and confirmed deletion of exon 5. (d) Conventional RT–PCR analysis confirmed expression of ARv5es transcript in D5 cells (upper panel). ARv5es is the major exon-skipping variant in D5 clones (lower panel). PCR products were individually subjected to DNA sequencing to further confirm that AR transcript in D5 is v5es at the nucleotide levels. (e) Total RNA was isolated from the specified cells treated with 100 μg/ml cycloheximide (CHX) for 7 h. H2O was used as a vehicle control (Ctrl). The primers P4 and P5 were used as described in Table 1 to assess AR-FL and v5es transcripts. Relative levels of AR and ATF1 transcripts were compared with the levels of the corresponding genes in 'Ctrl'-treated AD1 cell using RPL13A as a normalizer. All individual data points were plotted as the relative quantity (RQ). Similar results were obtained from three independent experiments. NMD inhibition uniformly increased ATF1 levels in all of the cell lines. ARv5es transcript responded much better to NMD inhibition than AR-FL counterpart.