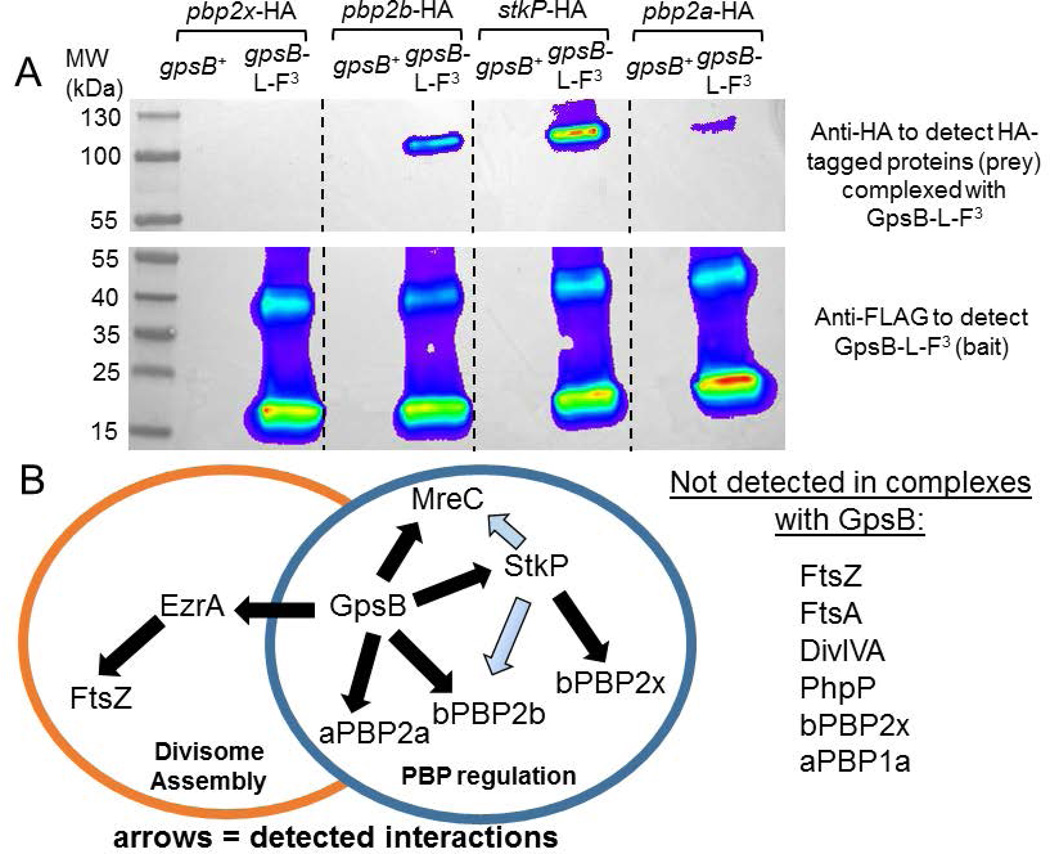
Fig. 9.
A) Pairwise co-IP of GpsB-L-FLAG3 with bPBP2b-HA, StkP-HA, or aPBP2a-HA4, but not with bPBP2x-HA. Co-IP experiments were performed as described in Experimental procedures. Top blot was probed with anti-HA primary antibody for HA-tagged prey proteins, using GpsB-L-FLAG3 as bait protein. 57 µg of each lysate sample were loaded on the input gel, while 20 µL of each elution sample was loaded on to the elution gel, after mixing 1:1 with 2× Laemlli sample buffer. Predicted molecular weight (MW) of bPBP2x-HA, bPBP2b-HA, StkP-HA, and aPBP2a-HA4 are 83.5 kDa, 75.7 kDa, 73.5 kDa, and 85.2 kDa, respectively. Bottom blot was probed with anti-FLAG primary antibody for GpsB-L-FLAG3 (bait). Two major bands are detected by anti-FLAG primary antibody in strains expressing GpsB-L-FLAG3. The bottom band correlates to GpsB-L-FLAG3 monomer (≈ 16.4 kDa), whereas the top band is likely a GpsB-L-FLAG3 trimer based on MW. Lanes shown on blot are as follows (all strains were constructed in the D39 Δcps background, IU1945): lane 1, pbp2x-HA gpsB+ (IU6929); lane 2, gpsB-L-FLAG3 pbp2x-HA (IU11314); lane 3, pbp2b-HA gpsB+ (IU6933); lane 4, gpsB-L-FLAG3 pbp2b-HA (IU11316); lane 5, stkP-HA gpsB+ (IU7438); lane 6, gpsB-L-FLAG3 stkP-HA (IU11412); lane 7, pbp2a-HA4 gpsB+ (IU11560); and lane 8 pbp2a-HA4 gpsB-L-FLAG3 (IU11516). This experiment was performed twice with similar results. B) Map of interactions found by in vivo co-IP that are proposed to coordinate divisome assembly with PBP regulation. GpsB was detected in complexes with EzrA, StkP, aPBP2a, bPBP2b, and/or MreC at stages of the division cycle (above; Fig. S10, S11, and S14). StkP was detected in complexes with bPBP2x, bPBP2b, and MreC (Fig. S13 and S14), although complexes with bPBP2b and MreC could be indirect (blue arrows) via interactions of these proteins with GpsB. EzrA is in complexes with FtsZ and GpsB (Fig. S11 and S12) and other division proteins not shown (Amilcar Perez, in preparation for submission). GpsB did not pull down detectable levels of FtsZ, FtsA, DivIVA, PhpP, bPBP2x, or aPBP1a by this in vivo co-IP method (above; Fig. S10–S12 and S14).