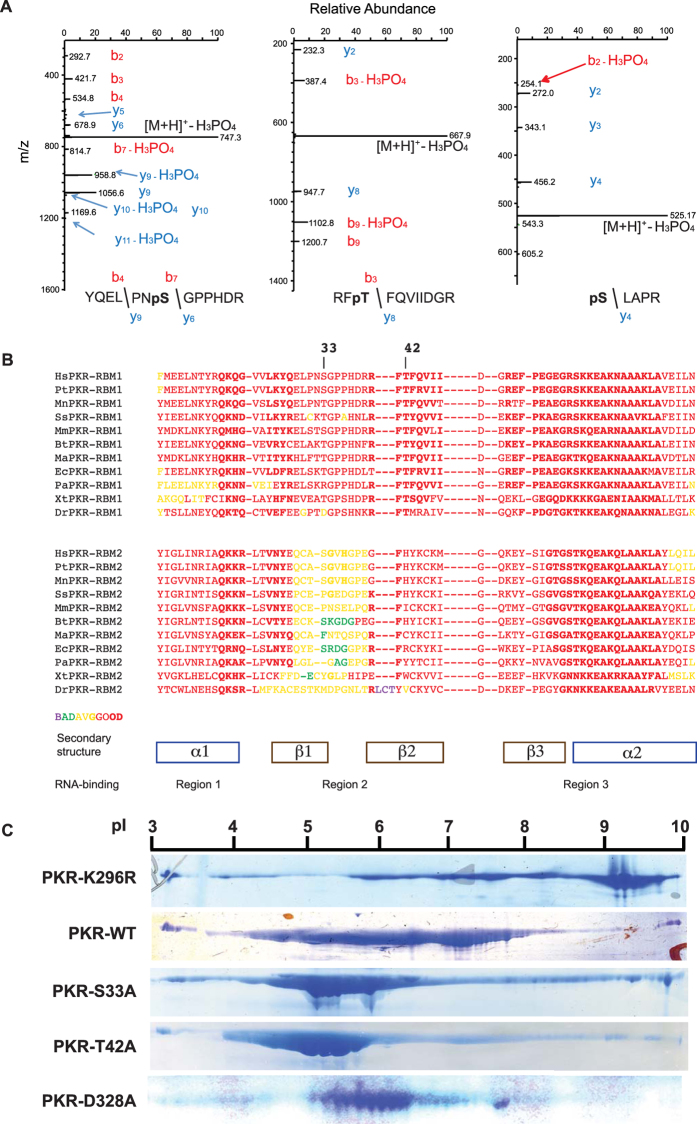
Figure 1. Phosphor-residues in RBM1 repress auto-phosphorylation of PKR.
(A) LC-MS/MS analysis of the tryptic digest of PKR identifying phosphor-S33 and -T42 residues as doubly and phosphor-S242 as singly charged peptides, with m/z ratios of 795, 717 and 623 Da, respectively. The fragmentation spectra of the peptides are dominated by loss of H3PO4 from the precursor ion, which is indicative of the presence of a pS or pT residue and the mass of this ion is consistent with the YQELPNSGPPHDR, RFTFQVIIDGR or SLAPR + PO3 peptides. The site of modification is evident by the mass of the: N-terminal b4 and b7 ions, which is consistent with modification on S33 (YQELPNpSGPPHDR); N-terminal b3 ion, which is consistent with modification on T42 (RFpTFQVIIDGR); and C-terminal y4 ion, which is consistent with modification on S242 (pSLAPR), as indicated. (B) An alignment of the amino acid sequences of the PKR RBM1 (above) and RBM2 (below) from the indicated species; Hs = Homo sapiens, Pt = Pan troglodytes, Mn = Macaca nemestrina, Ss = Sus scrofa, Mm = Mus musculus, Bt = Bos taurus, Ma = Mesocricetus auratus, Ec = Equus caballus, Pa = Pteropus alecto, Xt = Xenopus tropicalis, Dr = Danio rerio. A more extensive alignment to RBMs from proteins other than PKR is shown in Supplementary Figure S1. (C) The indicated recombinant PKR proteins electrophoretically separated by their isoelectric point (pI), then visualized with the Coomassie brilliant blue stain.