Figure 6. Phosphor-S33 and -T42 residues control the association between RBM1 and the kinase domain and PKR dimerization.
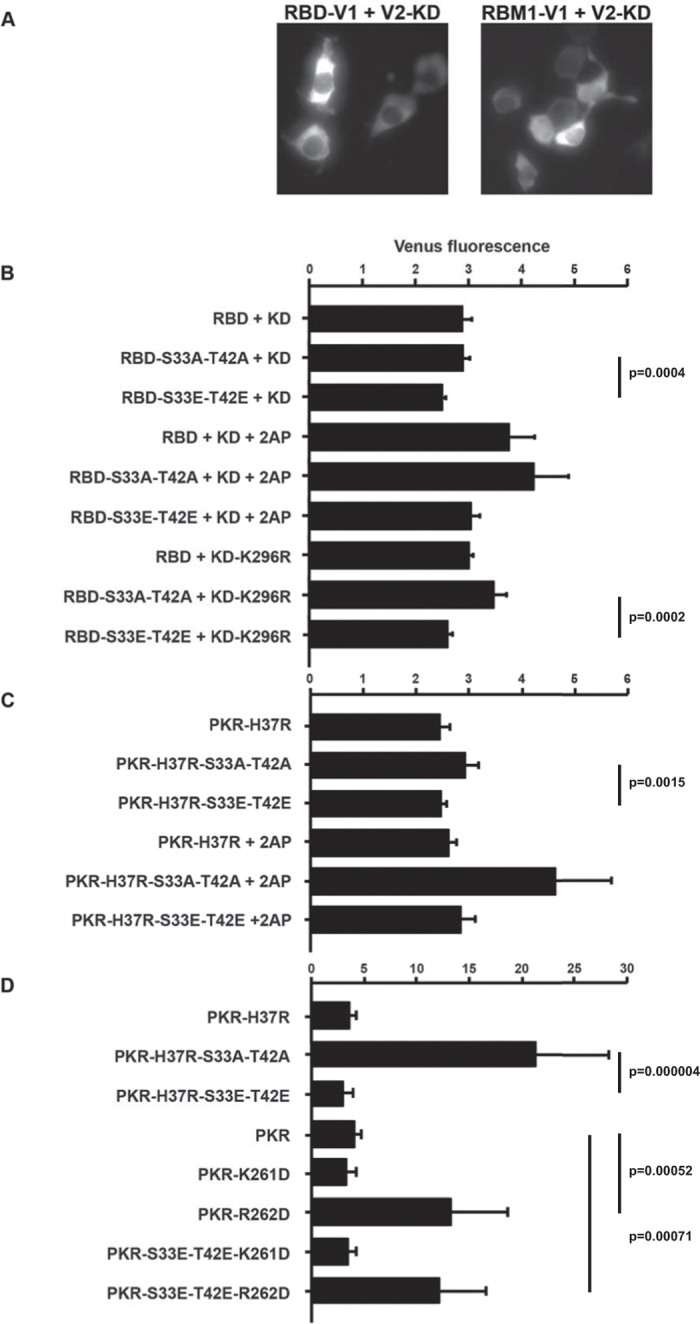
(A) Micrographs showing Venus fluorescence produced by peptide association in HEK293 cells expressing the RNA-binding domain (RBD) or just RBM1 and the kinase domain (KD) with split Venus as C-terminal V1 and N-terminal V2 tags, respectively (using 20 ng/well of each plasmid construct). (B) A quantitation of Venus fluorescence produced by an association between the indicated RNA-binding domain (RBD) constructs and the truncated kinase domain (KD) of PKR tagged with separate halves of the split Venus fluorophore as C-terminal V1 or N-terminal V2 tags, respectively (using 20 ng/well of each plasmid construct). The effect of kinase activity was examined by treatment with 5 μM of the inhibitor 2-aminopurine (2AP) and by using the phosphor-transfer mutant kinase domain (KD-K296R) (n = 7 for the cells expressing the wild-type KD, n = 6 for 2AP-treated cells and n = 5 for experiments with the KD-K296R construct). (C) A quantitation of Venus fluorescence produced by dimerization of the indicated full-length PKR constructs and the truncated kinase domain tagged with separate halves of the split Venus fluorophore as N-terminal V1 or V2 tags, respectively (using 20 ng/well of each construct). The histidine residue number 37 was mutated to an arginine (H37R) to reduce RNA-binding to RBM1. The effect of kinase activity was examined by treating transfected cells with 5 μM of 2AP (n = 7 for the cells untreated and n = 3 for cells treated with 2AP). (D) A quantitation of Venus fluorescence produced by dimerization of the indicated full-length PKR constructs and the truncated phosphor-transfer mutant kinase domain (KD-K296R) with N-terminal V1 or V2 tags, respectively (using 20 ng/well of full-length PKR and 30 ng/well of the truncated KD). The lysine and arginine resides number 261 and 262, respectively, within the α0-helix of the kinase domain, are mutated to aspartic acids (K261D and R262D) to test the involvement of either residue in the interaction with the 33 and 42 residues in RBM1 (n = 7). Error bars show the SEM in all graphs. Student’s t test was used to calculate the p values.
