FIGURE 4.
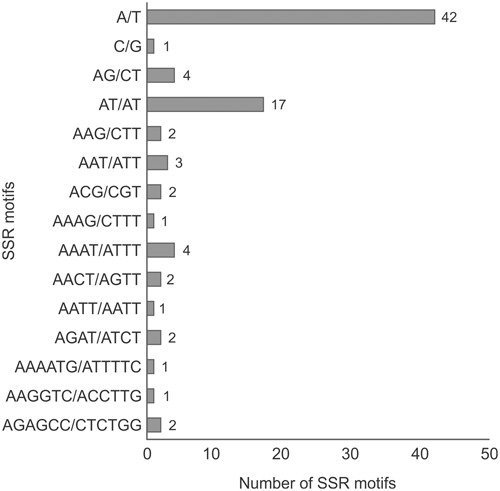
The number of SSR motifs found in the P. edulis chloroplast genome, taking into account sequence complementarities. The criteria used to search SSR motifs were set as follows: motifs between one and six nucleotides long, with a minimum repeat number defined as 10, 5, and 4 units for mono-, di-, and trinucleotide SSRs, respectively, and three units for each tetra-, penta-, and hexanucleotide SSRs.
