Fig. 2.
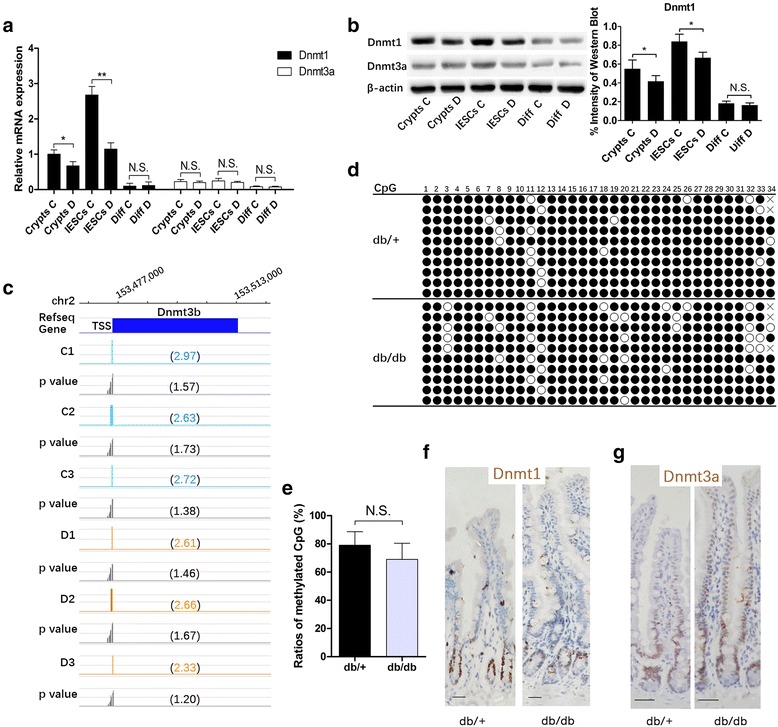
Expression and promoter methylation of DNA methyltransferases in intestinal epithelium. a, b Decreased Dnmt1 expression is detected in db/db crypts and intestinal epithelium stem cells (IESCs) by qRT-PCR using the FACS cell population (a) and Western blot using the MACS cell population (b left) and quantification of immunoblot bands from Western blot (b right). mRNA and protein levels are expressed relative to β-actin. Mean ± SE; n ≥ 6; *P < 0.05, **P < 0.01 by t test. C represents the control db/+ group; D represents the diabetic db/db group. c Promoter methylation microarray of Dnmt3b indicates promoter hypermethylation not only in db/db mice (orange bars) but also in db/+ mice (light blue bars). Values in brackets indicate the peak score that reflects the probability of positive enrichment and average P value scores. d, e Validation of Dnmt3b microarray results by bisulfite sequence. The table indicates the results of bisulfite sequencing analyses of 10 bacterial colonies per group (d). The CpG sites validated are the sites that exhibit a peak signal in the microarray. Black circles represent methylated CpG sites; white hollow circles represent unmethylated CpG sites; crosses represents methylation status not determined . Bar graph showing the ratio of methylated cytosine in CpG within the regions analyzed (e). f, g Immunohistochemical staining of Dnmt1 (f) and Dnmt3a (g). Scale bars = 50 μm. Diff differentiated cells, N.S. not significant
