Fig. 1.
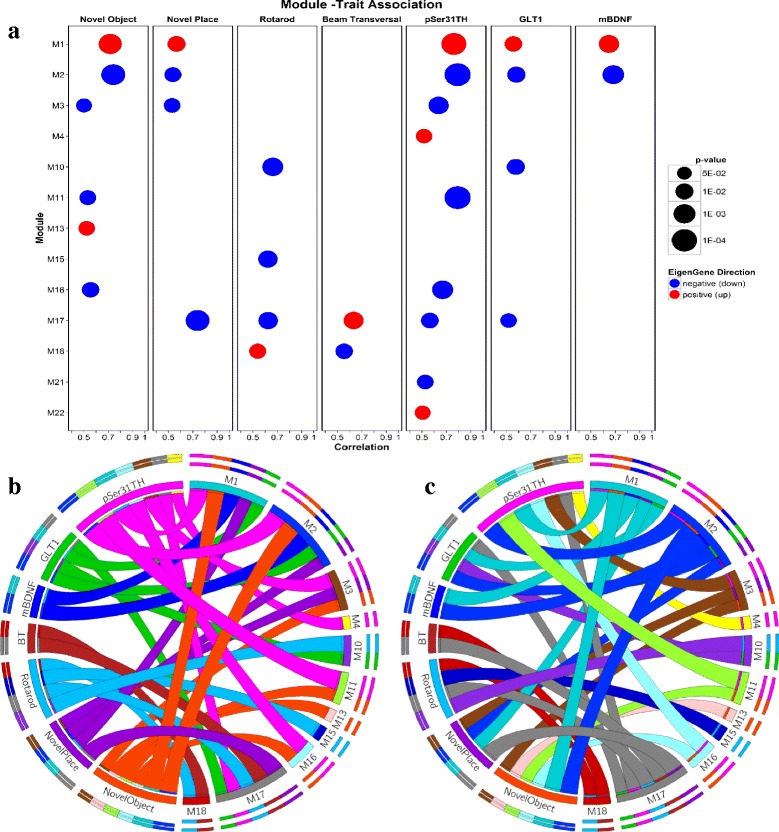
Module-Trait Association. a All 31 significant associations between 13 modules and 7 phenotypes are depicted as determined by bi-weight midcorrelation (bicor) between the first principal component (or EigenGene, ME) of gene expression and the continuous phenotypes (p < 0.05, absolute value of r > 0.5). Each circle describes a single significant correlation related to the module in association to a phenotype. The size of the circle corresponds to the significance of p-value. Circle color signifies the direction of the correlations and of MEs (blue = upregulated, red = downregulated). In both panels (b) and (c), the left hemisphere of the Circos plot represents the quantitative phenotypes in color-coded segments respectively while the right hemisphere represents the significant modules (M) displayed in color-coded segment designated by WGCNA. Each ribbon denotes a significant correlation between a phenotype and a module (29 significant associations). The number of the ribbons originating from each segment’s inner rim indicates the number of significant correlations between a phenotype and modules. Individual ribbon width demonstrates the strength of association calculated by bicor. Panel (b) demonstrates significant correlations colored by the origin of the phenotypes whereas panel (c) demonstrates significant correlations colored by the origin of the modules
