Fig. 1.
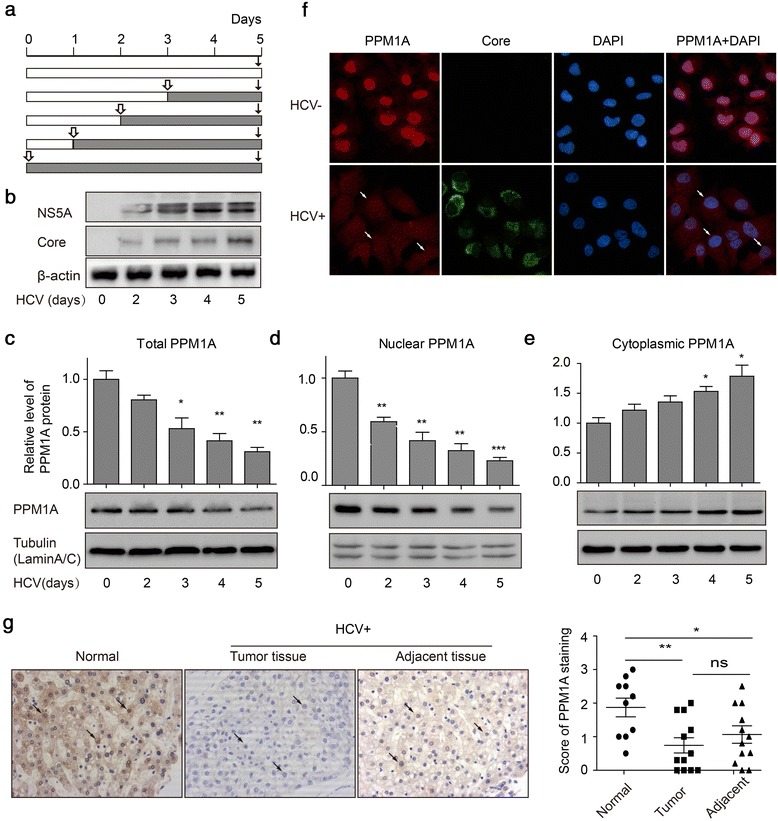
PPM1A abundance is negatively regulated in HCV-infected cells. a Schematic representation of the experiments shown in (b–e). Huh-7 cells were infected with JFH1 virus for 0–5 days at 1 multiplicity of infection (MOI) at intervals, and subsequently lysed simultaneously for immunoblot analysis. Hollow bars show uninfected cells; shaded bars, infected cells. b Levels of HCV infection in Huh-7 cells were determined by immunoblotting for HCV NS5A and core protein as described in a. c–e Total (c), nuclear (d), and cytoplasmic (e) PPM1A protein levels were measured by western blotting. The bar graph (mean ± SEM) displays protein quantification (n = 3). Data are normalized to a loading control (tubulin for total and cytoplasmic PPM1A, and lamin A/C for nuclear PPM1A) and expressed as the fold change relative to the protein levels in uninfected cells. f Cells were infected or not with JFH1 for 3 days, and the subcellular localization of PPM1A was monitored by dual immunolabeling of HCV core protein (green) and PPM1A (red). Arrows denote HCV-infected cells in which PPM1A expression and subcellular localization are significantly changed. g Immunohistochemistry was used to measure the expression of PPM1A (brown staining) in normal liver tissues (n = 10), and HCV-related HCC tissues and adjacent tissues (n = 12 each). g Immunohistochemistry was used to measure the expression of PPM1A (brown staining) in normal liver tissues (n = 10), and HCV-related HCC tissues and adjacent tissues (n = 12 each). Arrows indicate representative staining of PPM1A. Left panels show representative images of PPM1A expression, quantitative data are shown in the right panel. Data are the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 as evaluated using Student’s t-test or one-way ANOVA
