Fig. 4.
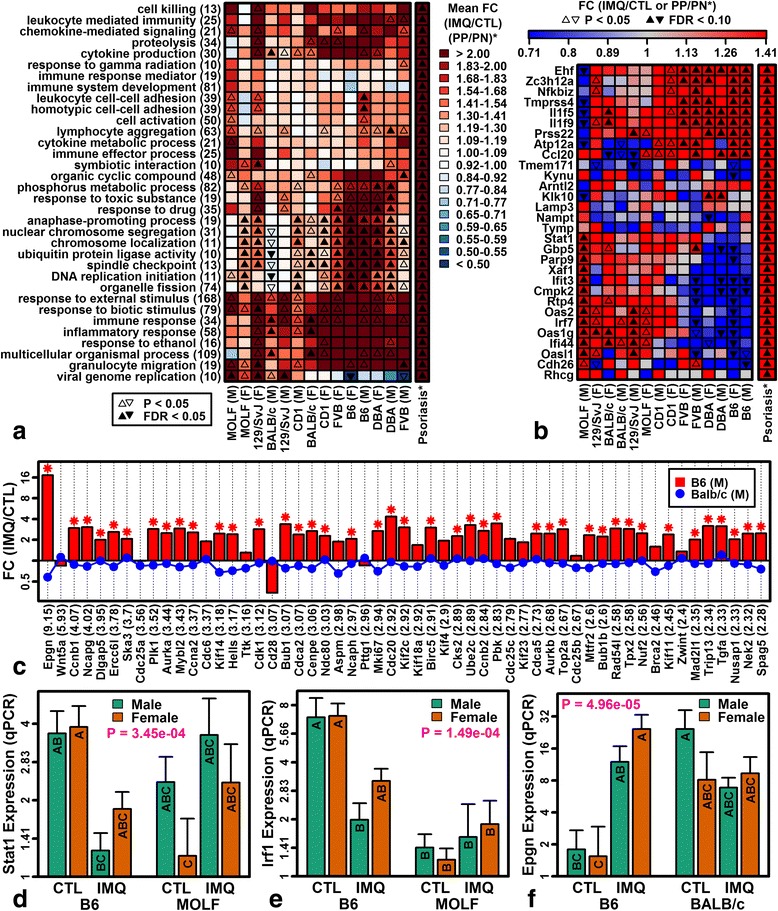
Gene Ontology (GO) biological process (BP) terms enriched among psoriasis-increased DEGs and correspondence with IMQ responses. a Psoriasis-enriched GO BP terms. The heatmap shows average IMQ responses of ten representative mouse homologs for each GO BP term. Chosen genes are homologous to the ten human genes associated with the listed term for which expression was most strongly elevated in psoriasis lesions (i.e., lowest p value). b Psoriasis-increased DEGs induced by poly(I:C) in cultured human KCs. c Genes associated with “organelle fission” (GO:0048285) (*FDR <0.10, IMQ versus CTL). d–f RT-PCR analysis of Stat1 (d), Irf1 (e), and Epgn (f) expression (n ≥ 4 per strain/sex/treatment group; n = 38–40 mice total). Groups without the same letter differ significantly (P < 0.05, Tukey honest significant difference. Error bars represent standard error of the mean; p values strain-by-treatment interaction effect). Rn18s was used as an endogenous control to estimate relative gene expression. F female, FC fold change, M male, PN uninvolved skin from psoriasis patients, PP lesional skin from psoriasis patients, qPCR quantitative PCR
