Fig. (1).
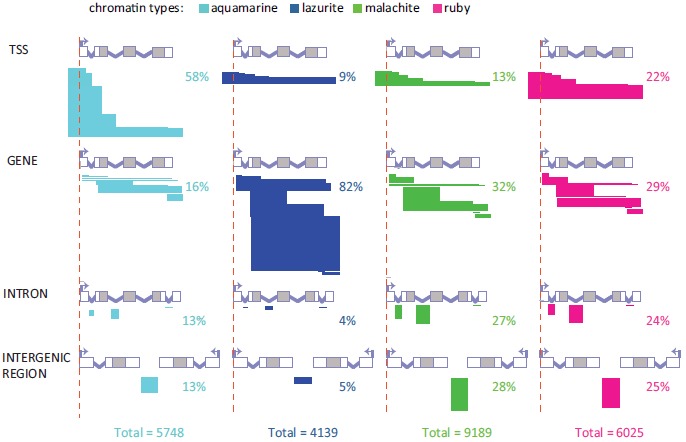
Detailed classification of the relative positioning of aquamarine, lazurite, malachite and ruby chromatin domains versus gene structure. Bent arrows and red vertical dashed lines indicate the positions of TSSs. Grey boxes represent coding parts of exons and white boxes correspond to 5’ and 3’ UTRs. Introns are shown as broken lines. Horizontal bars depict different possible overlaps between chromatin domains and genes for the all defined localization classes: TSS, GENE, INTRON and INTERGENIC REGION. The width of the bars reflects the percentage of each localization subclass in each chromatin type. For each localization class, fragments with borders mapped to various structural parts of a gene are shown separately.
