Figure 1.
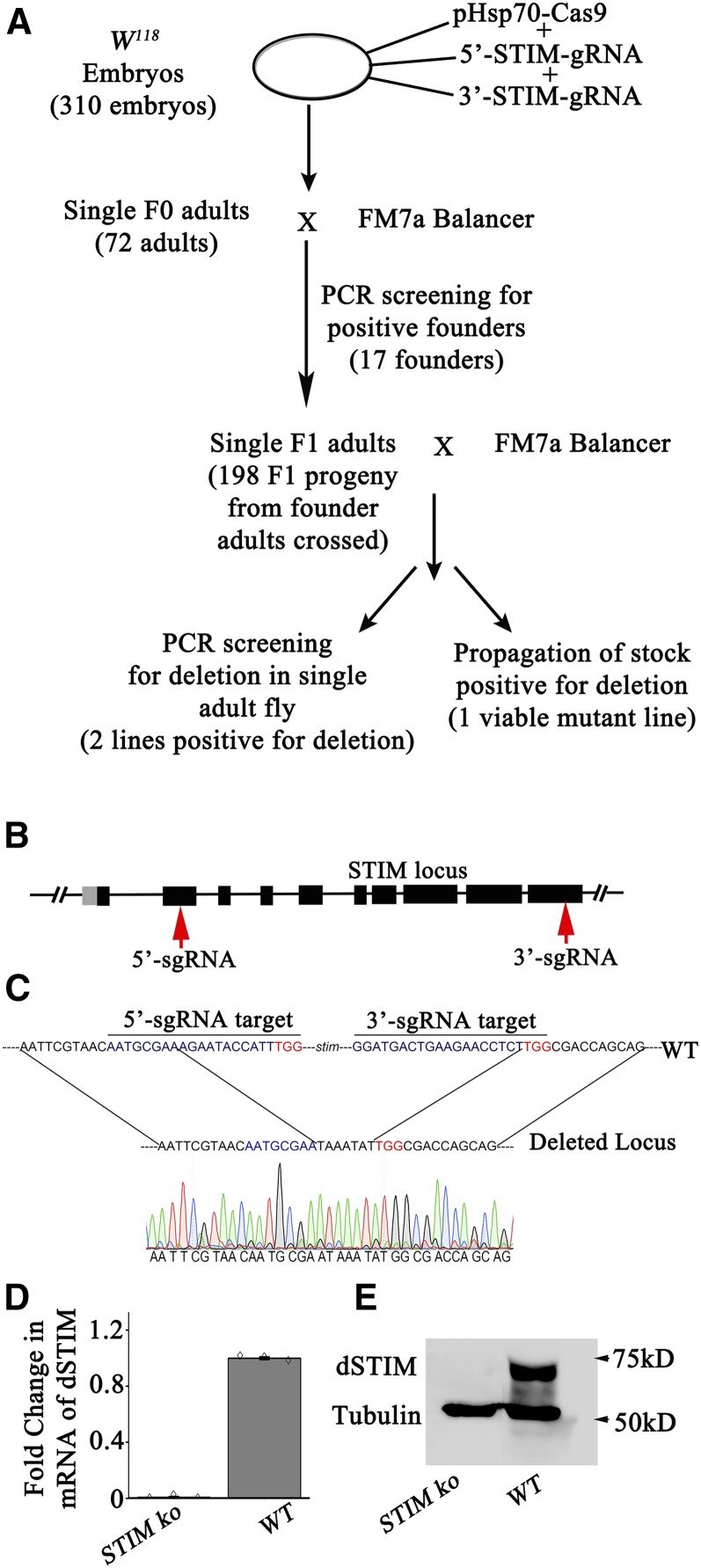
Knocking out dSTIM with the CRISPR-Cas9 system resulted in deletion of the dSTIM gene (A) Schematic representation for generation of STIM knockout (STIMko). Putative alleles were screened by PCR and balanced using first chromosome balancer FM7a. (B) Representation of dSTIM gene with exons (thick lines), introns (thin lines), and 5′−UTR (gray line). Target regions of guide RNAs are indicated with red arrows. (C) Sequencing of the dSTIM gene region confirming the deletion. (D) q-PCR of RNA isolated from STIMko second instar larvae (n = 3). The error bars represent SEMs. (E) Western blot of protein lysates from second instar larva of STIMko organisms. CRISPR, clustered regularly interspaced short palindromic repeats; PCR, polymerase chain reaction; q-PCR, quantitative PCR; UTR, untranslated region; WT, wild-type.