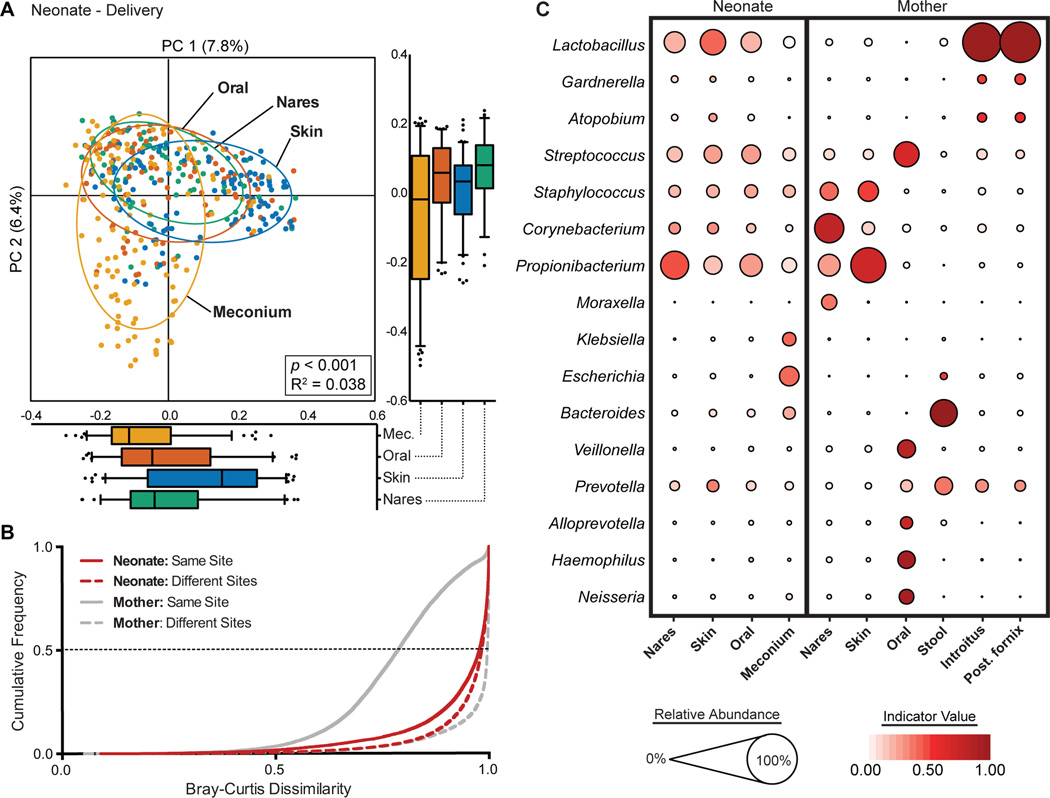
Fig. 1. Neonatal (at birth) microbial community structure.
(A) Principal coordinate analysis (PCoA) on unweighted UniFrac distances between the neonatal microbiota is shown along the first two principal coordinate (PC) axes. Boxplots shown along each PC axis represents the distribution of samples along the given axis, representing the median and interquartile range with whiskers determined by Tukey’s method. Each point represents a single sample and is colored by body site: Meconium (Mec.), Orange; Skin, blue; Oral Cavity, red; Nares, green. Ellipses represent a 95% confidence interval around the cluster centroid. Clustering significance by virtue of body site was determined by Adonis (p<0.001). (B) Cumulative distribution of Bray-Curtis dissimilarity distances calculated pairwise between samples of the same body site (solid lines), and between different body sites (dashed lines). Distance comparisons for neonates and maternal samples are shown in red and gray, respectively. Smaller values indicate a greater similarity between samples. (C) The average relative abundance (circle size) of the most prevalent genera (y-axis) in each body site (x-axis) is plotted for neonates and mothers at delivery. The indicator value index (related to circle color shade darkness) represents the strength of association between a taxa and a given body site, with larger values indicating greater specificity.