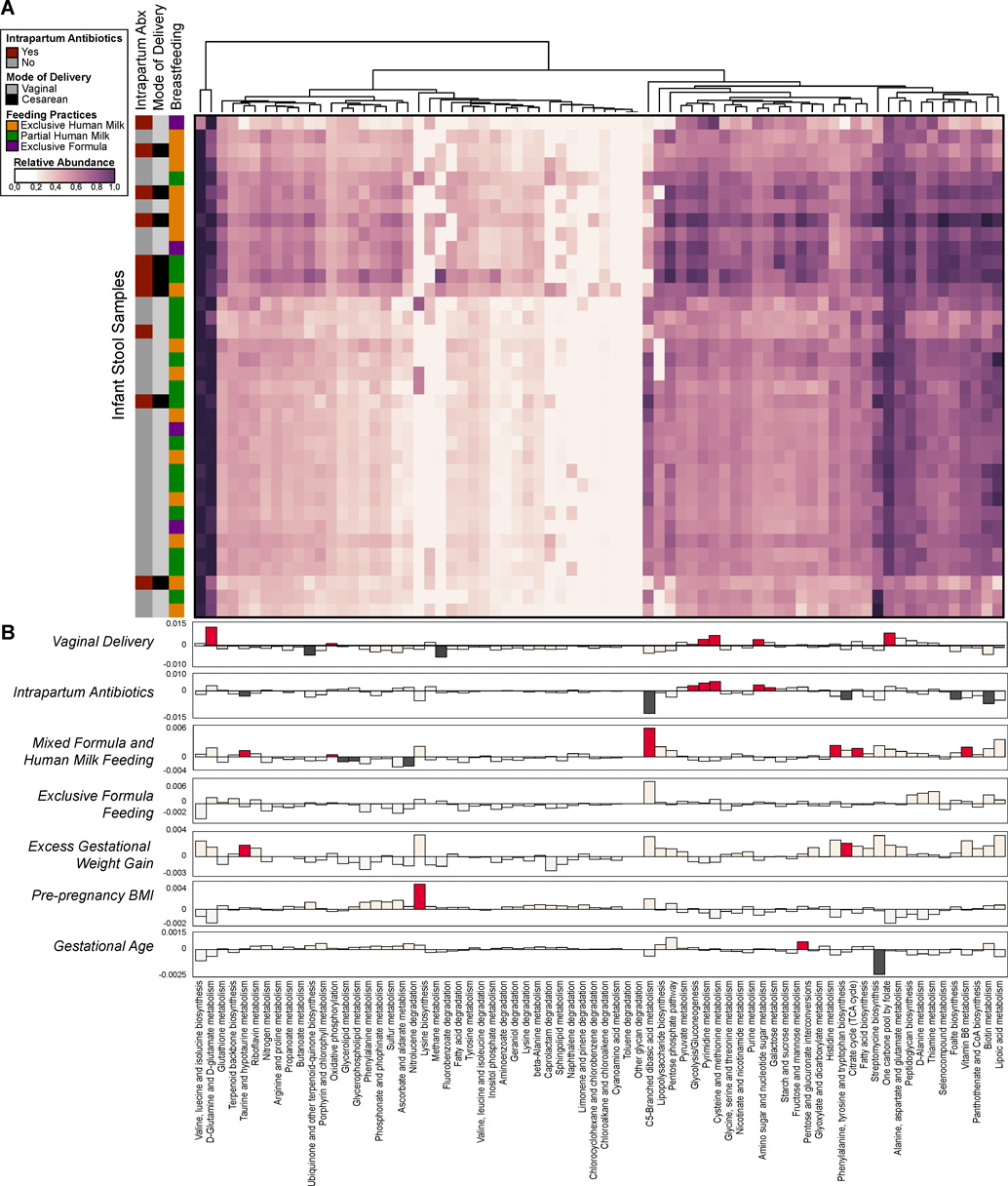
Fig. 6. Infant microbial community function with clinical metadata in a generalized linear model.
(A) Heatmap of the relative abundances of KEGG pathways found in infant stool samples as determined by WGS sequencing. The vertical bar on the left indicates mode of delivery, breastfeeding practices and intrapartum antibiotic usage. Dendrograms represent hierarchical clustering on Euclidean distances using average linkage. (B) A generalized linear mixed model was fitted for each pathway to identify pathways whose abundances differed significantly between individuals by virtue of mode of delivery, antibiotic usage, breastfeeding, gestational weight gain, BMI and gestational age. The strength of the linear model predictions for each pathway is represented by bar height. Significant correlations are indicated by the darker color (dark red or dark gray). Labels correspond to the following comparisons: Vaginal Delivery - pathways enriched in infants born vaginally (red, up) or by Cesarean (gray, down); Intrapartum Antibiotics - pathways enriched in infants exposed to intrapartum antibiotic usage (red, up) as opposed to no antibiotics (gray, down); Mixed Formula and Human Milk Feeding - pathways increased in partially breast fed (human milk and formula) infants (red, up) as opposed to exclusive human milk only (gray, down); Exclusive Formula Feeding - pathways higher in exclusively formula fed infants (red, up), as opposed to human milk only (gray, down); Excess Gestational Weight Gain - pathways higher in cases of excess maternal gestational weight gain (red, up), as opposed to normal weight gain (gray, down); Pre-pregnancy BMI & Gestational age - pathways positively (up, red) or negatively (down, gray) correlated with pre-pregnancy BMI or gestational age.