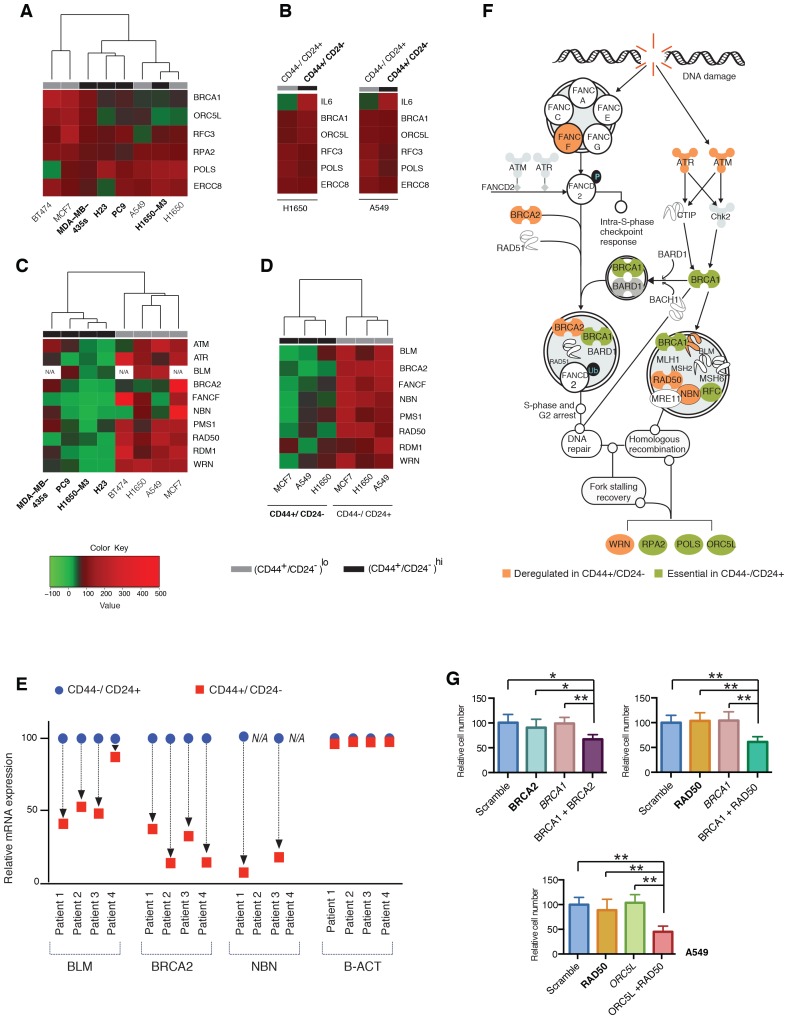
Figure 2. Decreased expression of homology-directed repair (HDR) genes in CD44+/CD24− cells results in synthetic lethal interactions.
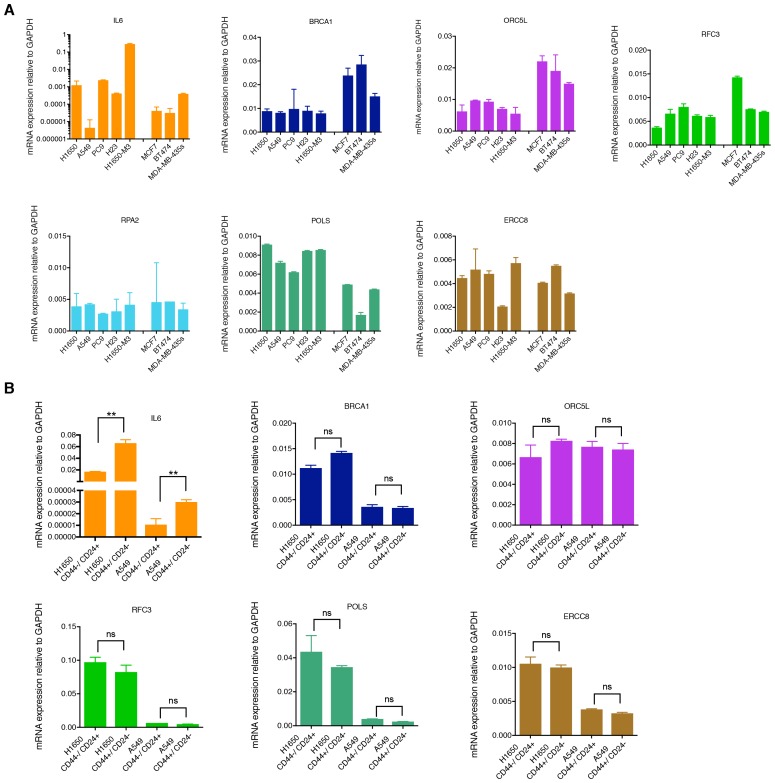
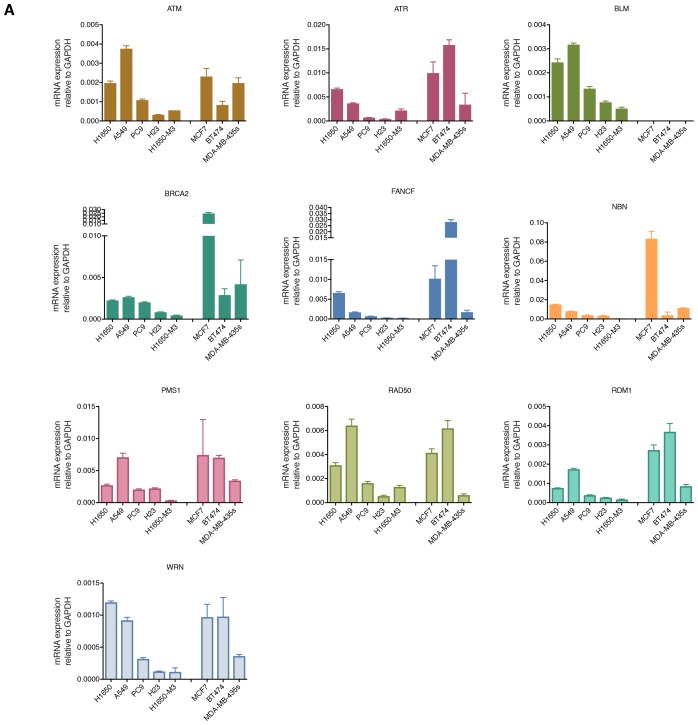
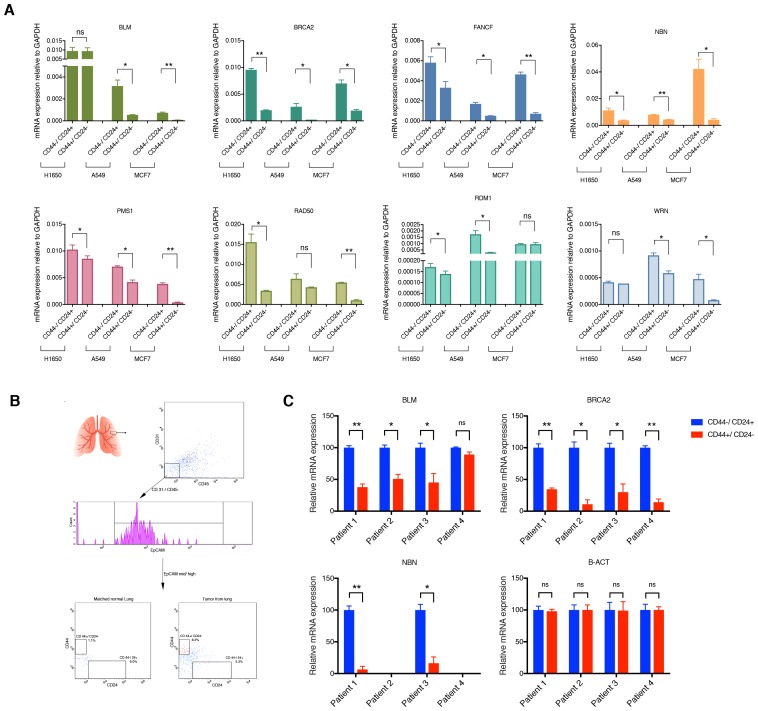
(A) The heat map represents a hierarchical cluster analysis of BRCA1, ORC5L, RFC3, RPA2, POLS and ERCC8 mRNA expression in the indicated tumor-derived cell lines and (B) in H1650 and A549 cells that were FACS sorted on the basis of their surface expression of CD44 and CD24. mRNA expression was quantified by SYBR-green-based RT-qPCR. Cell lines with high CD44+/CD24− cell content are indicated in bold. The data represent mean ± SD of three replicates from two independent experiments. See Figure 2—figure supplement 1 for details. (C) Hierarchical cluster analysis of the mRNA expression of the indicated HDR genes in multiple tumor-derived cell lines with high (indicated in bold) or low content of CD44+/CD24− cells. mRNA expression was quantified by SYBR-green-based RT-qPCR. The data represent the mean ± SD of three replicates from two independent experiments. See Figure 2—figure supplement 2 for details. (D) Clustering analysis of mRNA expression of the HDR genes in CD44+/CD24− and CD44−/CD24+ cells FACS-sorted from H1650, A549 and MCF7 cells. mRNA expression was quantified by SYBR-green-based RT-qPCR. The data represent the mean ± SD of three replicates from two independent experiments. See Figure 2—figure supplement 3A for details. (E) Expression of BLM, BRCA2 and NBN genes in FACS-sorted CD44−/CD24+ and CD44+/CD24− cells from four human primary NSCLC tumors. mRNA expression was quantified by SYBR-green-based RT-qPCR. Expression of an indicated mRNA in the CD44+/CD24− cells was calculated relative to its expression in CD44−/CD24+ cells from the respective tumor. Each dot represents mean ± SD of three replicates. See Figure 2—figure supplement 3B,C for details. (F) Schematic representation of the functional interactions between genes that we identified in the screen (green) and the HDR genes that we found to be downregulated (orange) in the H1650-M3 (and CD44+/CD24−) cells (see Supplementary file 2 for details). (G) The charts depict the percentage of viable cells 5 days after knockdown of the indicated genes relative to a scramble-siRNA control in the A549 cell line. Each bar represents mean ± SD of eight replicates from two independent experiments. See Figure 2—figure supplement 5 for knockdown efficiency (p-value *<0.05, **<0.005 paired t-test).