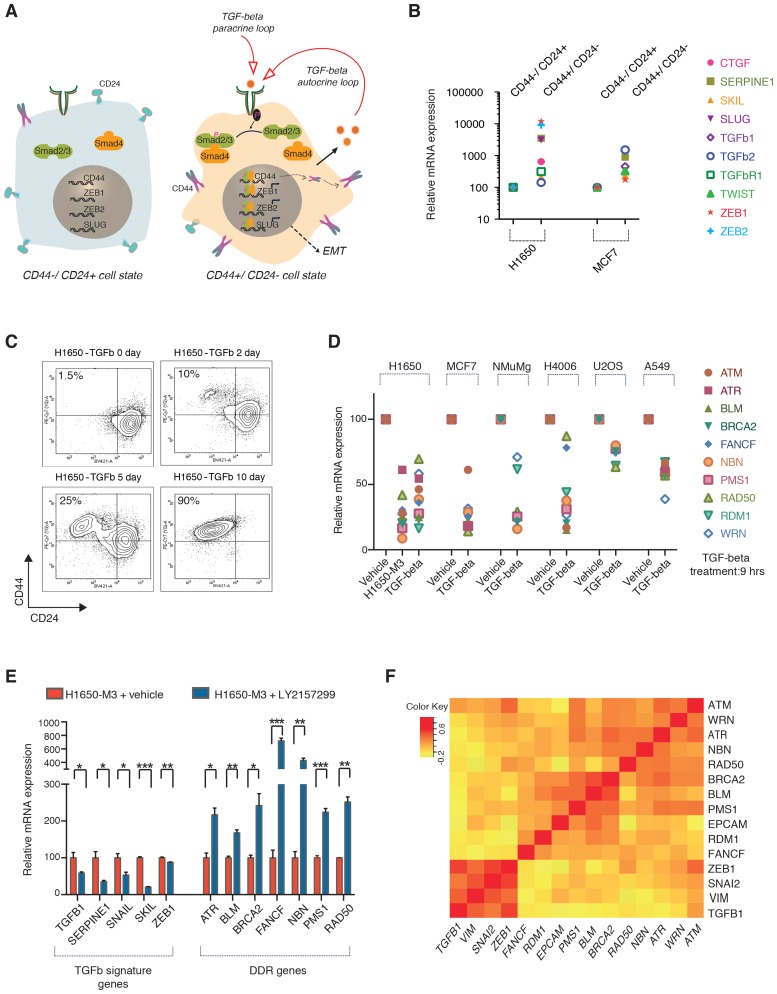
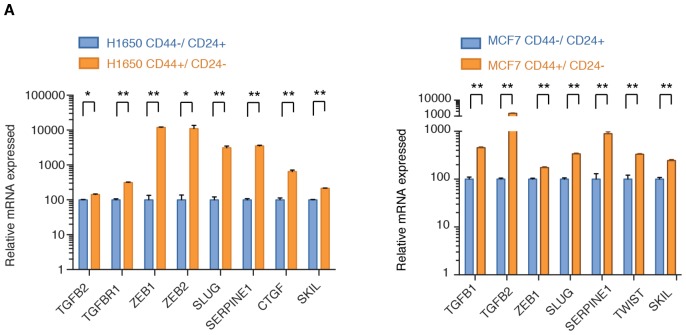
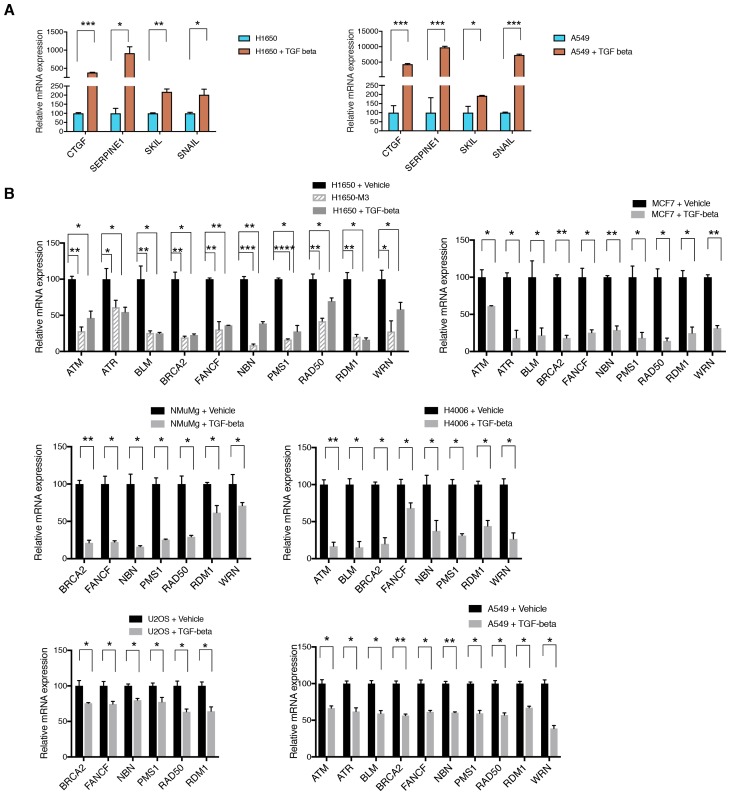
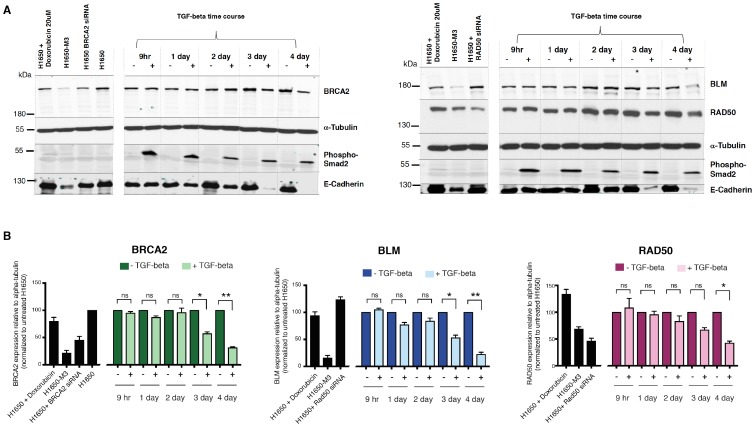
Figure 3. The TGF-β axis controls the expression of the HDR genes that are downregulated in CD44+/CD24− cells.
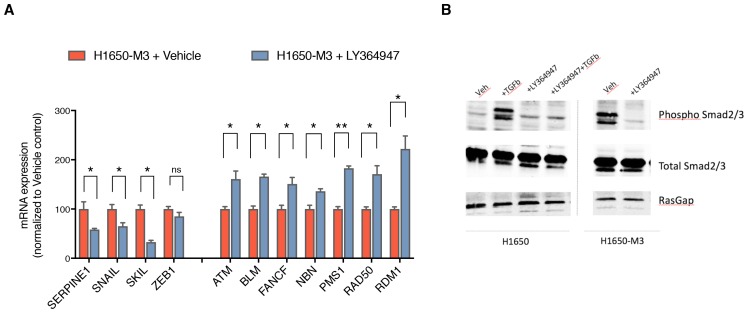
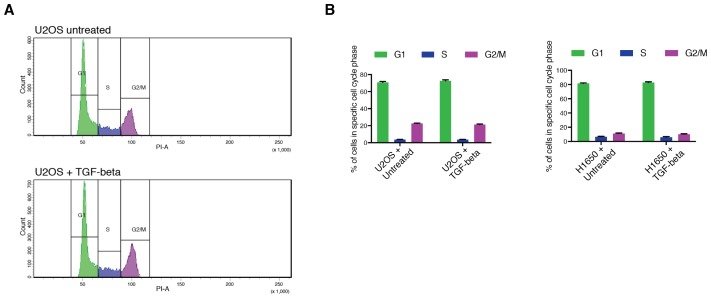
(A) TGF-β signaling is required for CD44+/CD24− cell state transition. The schematic provided here is based on current literature (Korkaya et al., 2011; Mani et al., 2008). (B) mRNA expression analysis of well-known TGF-β target genes in CD44+/CD24− cells relative to CD44−/ CD24+ cells FACS-sorted from H1650 and MCF7 cell lines. mRNA expression was quantified by SYBR-green-based RT-qPCR. Each dot represents the mean ± SD of three replicates from two independent experiments. See Figure 3—figure supplement 1 for details. (C) FACS analysis of H1650 cells exposed to TGF-β. Cells were treated with TGF-β for the indicated days and stained for the surface expression of CD44 and CD24 and analyzed by FACS. (D) mRNA expression analysis of the indicated HDR genes in TGF-β-treated cells relative to vehicle control across multiple tumor-derived cell lines. Cells were treated for 9 hr with TGF-β1 and TGF-β2 (1 ng/ml each). mRNA expression was quantified by SYBR-green-based RT-qPCR. Each dot represents the mean ± SD of three replicates from two independent experiments. See Figure 3—figure supplement 2B and Figure 3—figure supplement 3 for additional details. (E) The inhibition of TGF-β signaling in the CD44+/CD24− H1650-M3 cells results in an increased expression of HDR genes. TGF-β receptor 1 (TGFBR1) kinase activity was blocked by treatment with 20 μM of LY2157299 (Selleckchem) for 72–96 hr. Expression of TGF-β signature genes were used as a control for the efficacy of LY2157299 treatment. mRNA expression was quantified through SYBR-green-based RT-qPCR. Each bar represents the mean ± SD of three replicates from three independent experiments (p-value *<0.05, **<0.005 paired t-test). (F) Meta-analysis of human breast tumor dataset (BRCA) generated by the TCGA Research Network: http://cancergenome.nih.gov/. Average Pearson correlation coefficients (PCCs) between every pair of genes displayed were calculated and the matrix was generated. Pearson correlation coefficients ranged from −1 to +1. The matrix indicates an inverse co-regulation of the TGF-β1 and the HDR genes that we found to be downregulated in CD44+/CD24− cells.