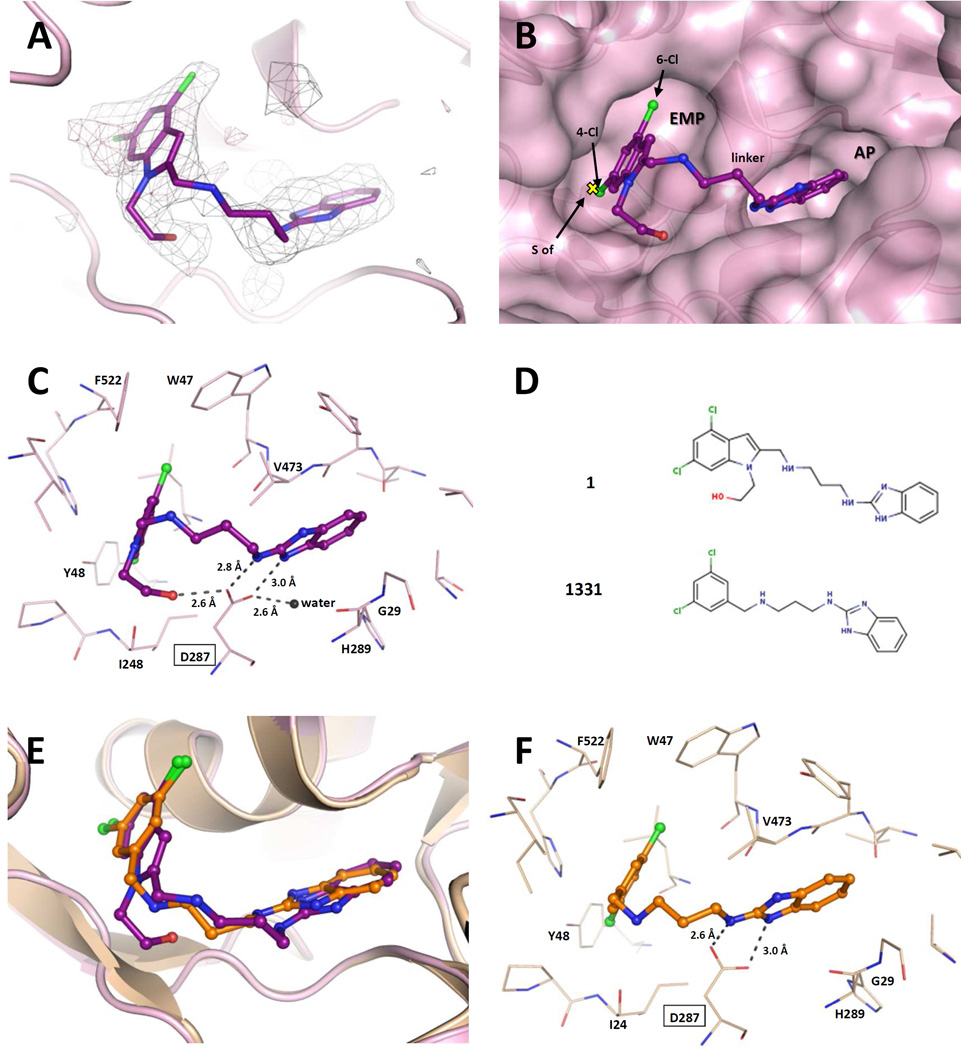
Figure 6.
Binding of 1 to TbruMetRS. (A) The TbruMetRS•1 structure (PDB: 5TQU) with the difference electron density map calculated by omitting the inhibitor, contoured at the 3σ level (positive density in grey, negative density in red). (B) General features of the 1 binding mode. The position of the methionine sulfur atom in the TbruMetRS•methionine structure (PDB: 4EG1)20 is shown with a yellow cross. The protein surface and the two pockets, EMP and AP, where the inhibitor is bound are shown. (C) Hydrogen bond network in the TbruMetRS•1 structure. The label for residue Asp287 is highlighted with a rectangle. The hydroxyethyl substituent of the 4,6-dichloroindolyl-ring moiety establishes an extra hydrogen bond with residue Asp287. (D) Chemical structures of compound 1 and the previously reported TbruMetRS inhibitor 1331. (E) Superposition of compounds 1 and 1331 (PDB: 4EG7)20 bound to TbruMetRS. (F) Hydrogen bond interactions between inhibitor and enzyme in the TbruMetRS•1331 structure.