Figure 2.
Can iesRNAs Be Generated from Ligated Excised IESs?
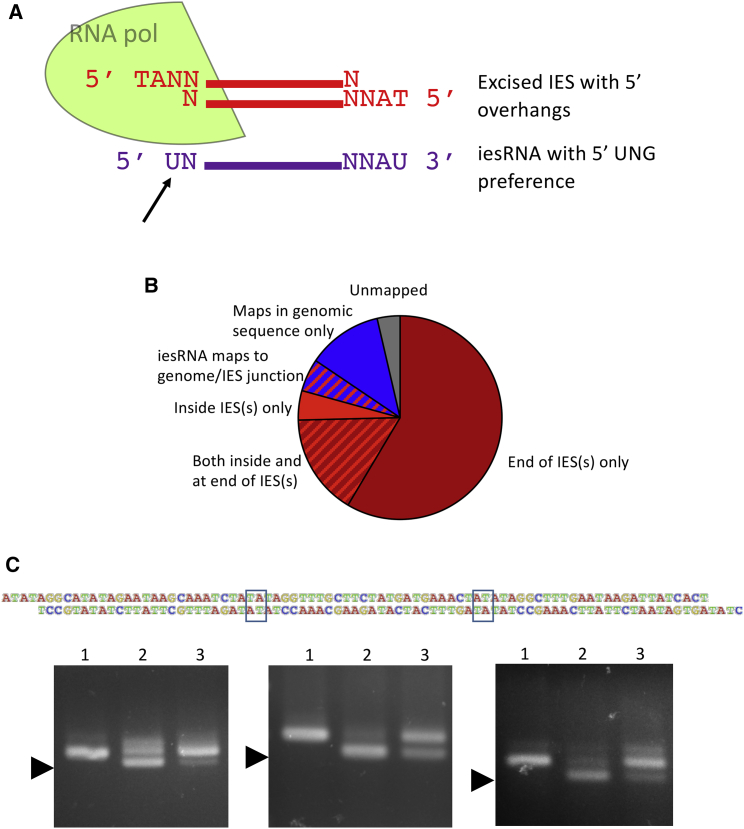
(A) Representation of the logistical problem of iesRNA transcription from excised IESs. A schematic excised dsDNA IES is shown in red, with its 4 nt 5′ overhangs. The corresponding iesRNA as deduced from small RNA mapping (Sandoval et al., 2014) is shown below in purple. The assembly of an RNA polymerase complex on a short (27 bp) DNA molecule is probably not possible. Even if it were, the RNA (purple) that corresponds to the 5′ end of the excised IES lacks a template for its 5′ nucleotide (indicated with a black arrow).
(B) Two hundred iesRNAs that map only partially to IES ends, with overhangs of at least 12 bp, were identified, and the overhangs were mapped. These iesRNAs were chosen randomly by the software Geneious 8.1.8. Overhangs were sorted into groups according to where they map. The majority map at the ends of other IESs, which is a huge overrepresentation considering that the number of possible mapping sites for a random 12 nt sequence within an IES is much greater than at its ends.
(C) The oligonucleotide corresponding to three ligated cryptic IESs is depicted, with the TAs that form the theoretical ligation points outlined. Gels of PCR products for each individual 27 bp cryptic IES (all on different chromosomes) are shown. Arrowheads indicate bands corresponding to excision products. Successful injection of the double-stranded oligonucleotide led to excision of all three cryptic IESs from the genome. This was successfully repeated in eight independent injected cells. All cells that exhibited excision of one cryptic IES following concatemer injection also exhibited excision of the other two.