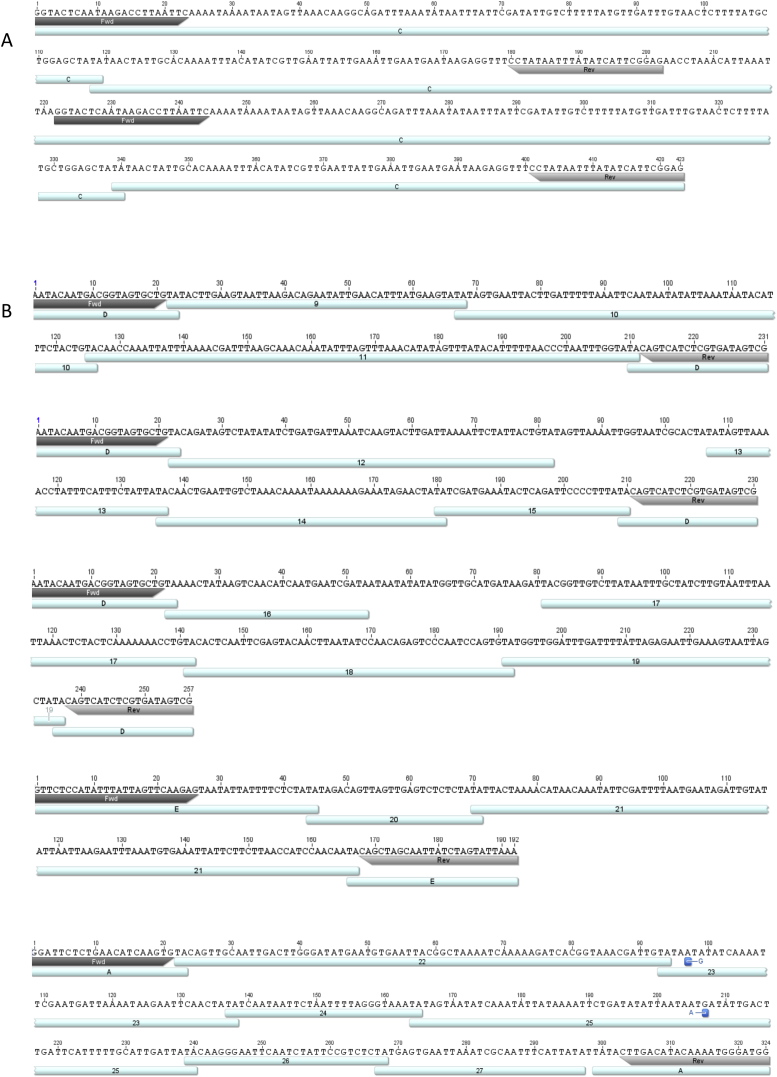
Figure S2.
RNA Transcripts of Concatenated IESs, Related to Figure 3
(A) RNA transcript of a repetitive IES sequence, presumably deriving from a circularised IES (> 200nt in length). Primer sites on the 221bp long IES are annotated, which amplify each IES end followed by the sequence of the opposite IES end. (B) Sequencing results of transcripts from concatenated IES. Primer binding site containing IESs are annotated in letters (A-E) and internal IESs in numbers (1-27). Gaps in sequence annotation result from lack of corresponding sequences in the database. Sequencing the Paramecium MIC genome is expected to reveal additional IESs that are not documented yet, but are present in these concatemers. iesRNAs exclusively map IESs and also map the gap sequences, suggesting these are non-annotated IESs. Point substitutions in the IES sequence are marked in blue and likely derive from sequence amplification with GoTaq® polymerase that lacks proofreading activity. In several cases, only partial IES sequences are detected, suggesting additional cleavage of these IESs occurs either while excision from the genome or after concatenation and circle formation. One example is IES C in A, where nucleotides 5-227 of the total 287bp long IES are forming the repeat.