Fig. 1.
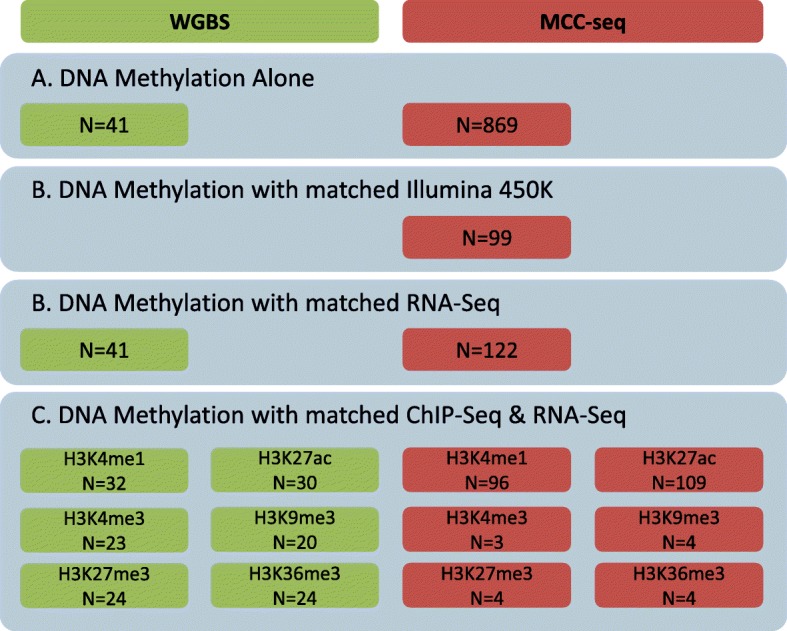
Samples having multiple layer of epigenetics profiles were used in this project. DNA methylation profiles were assessed for all samples using either whole genome bisulfite sequencing (WGBS; green) or targeted bisulfite sequencing (MCC-Seq; red). Listed in this figure are the number of samples used for analyses focusing on: a methylation sequencing alone (Methyl-Seq), b methylation with matched RNA-Seq from the same sample, or c methylation sequencing with matched ChIP-Seq (using six different histone marks) and matched RNA-Seq from the same sample
