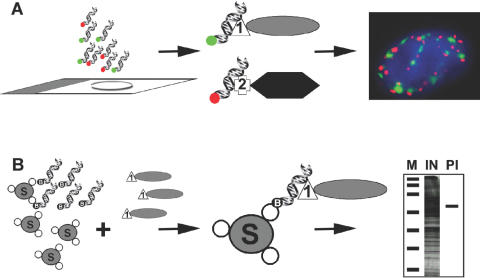
Figure 1.
In situ detection and isolation of proteins using dsDNA oligonucleotides. (A) Detection of proteins in situ using dsDNA oligos. Cells grown on slides are fixed and hybridized with fluorescently labelled dsDNA oligos (red or green) that can detect the presence of proteins fused to either LacI (1) or TetR (2). The fluorescent dsDNA oligos corresponding to the operator sequences for LacI or TetR (green or red, respectively) bind each bacterial fusion protein specifically without cross hybridizing resulting in the sub-nuclear localization of the tagged proteins by light microscopy. (B) Isolation and detection of proteins in vitro using dsDNA oligos. Streptavidin-sepharose beads (S) are sequentially incubated with biotinylated dsDNA oligos representing the operator sequences for LacI followed by incubation with a cell lysate containing LacI fused to a protein of interest (1). After binding the beads are washed and the resulting purified LacI-fusion protein can be observed by SDS–PAGE. M, marker, IN, input lysate, PI, isolated protein.