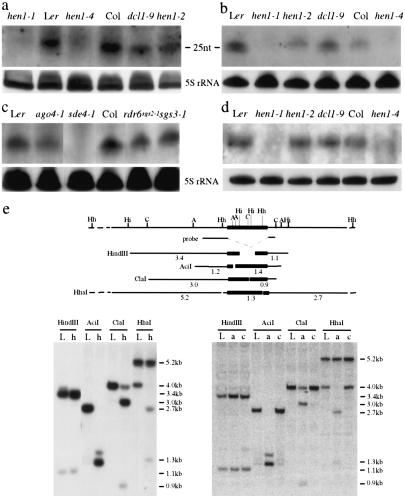
Figure 3.
Transposon siRNA accumulation and DNA methylation in various genotypes. Sense (a,c) or antisense (b) probes both detected siRNATEs. (d) Detection of siRNAs corresponding to AtSN1 in various genotypes. The 5S rRNA served as an internal control. rdr6sgs2-1, sgs3-1, and hen1-4 are in the Col accession; sde4-1 is in the C24 accession; and all other genotypes are in the Ler accession. The size of the siRNATEs was estimated with a 10-nt RNA ladder (Ambion) and by probing the same blot to visualize miR173 of known size (22 nt). (e) Determination of DNA methylation status at FLC-Ler in Ler (L), hen1-1 (h), ago4-1 (a), and cmt3-7 (c). A diagram of FLC-Ler genomic region is shown on top with the positions of restriction sites as indicated: (Hh) HhaI; (Hi) HindIII; (C) ClaI; (A) AciI; (Hh) HhaI. The probe corresponds to sequences flanking the TE (black box). The fragments that can hybridize to the probe in various digests are diagramed. The size of the fragments is indicated by numbers (in kilobases) underneath the fragments.