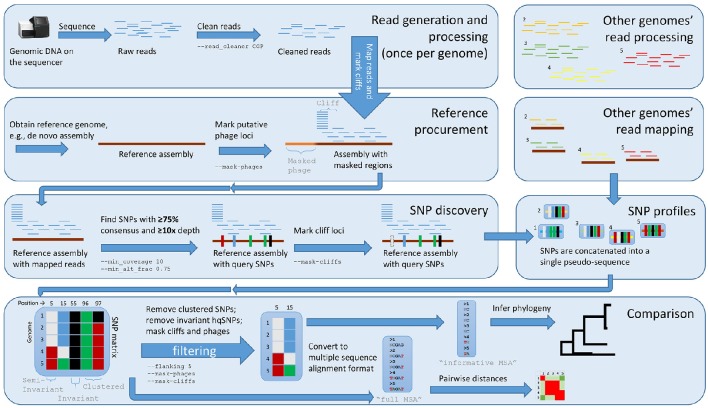
Figure 1.
The Lyve-SET workflow. Starting from the top left, reads are generated from a single query genome and then compared against a reference genome. Starting from the top right, other genomes are being generated and compared against the reference genome simultaneously. The order is (1) sequence query genome; (2) obtain a reference genome; (3) discover SNPs in a comparison against the reference genome; (4) combine SNP profiles into (5) a SNP matrix. In the bottom portion, the SNP matrix is interrogated for low-quality sites including those that are invariant or semi-invariant (those with masked or reference alleles). The matrix is also interrogated for clustered SNPs, i.e those that appear too close to each other. After the SNP matrix is queried and filtered, Lyve-SET obtains high-quality SNPs which are then used for creating a phylogeny. The larger, unfiltered multiple sequence alignment is used to calculate pairwise distances which can be used in a comparison, e.g., a heat map.