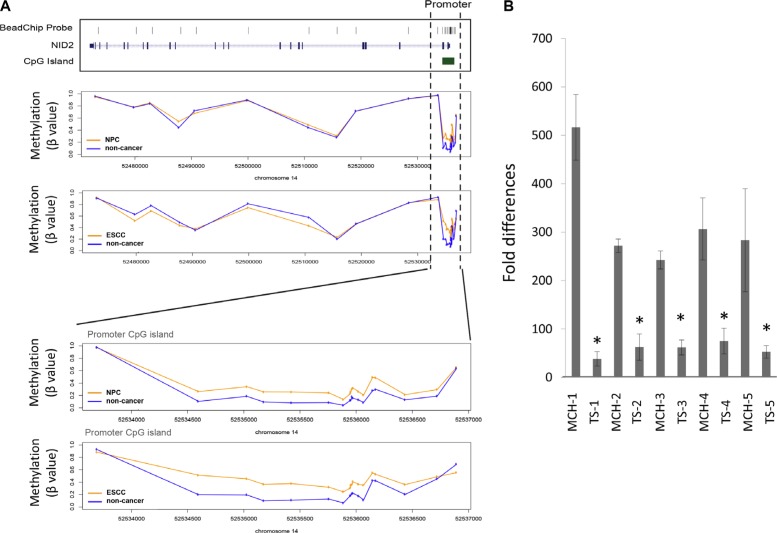
Figure 1. NID2 is identified as a candidate gene in NPC and ESCC.
(A) The average methylation level of NID2 derived from our previous methylome data in NPC and ESCC. The vertical broken line shows the region covering the promoter CpG island (chr14: 52534582–52536722) and the bottom figures show a close-up view of changes in methylation. Methylation level is presented as β value (β = M/(U+M+100), M: signal intensity of the methylated allele, U: signal intensity of the unmethylated allele). The y-axis shows the average methylation level in tumors (orange line) and non-cancer controls (blue line), respectively. Within this region, the methylation levels in multiple CpG sites of both NPC and ESCC patients are consistently higher than those of non-cancer controls, with adjusted p value < 0.05 estimated by LIMMA analysis using the transformed β values as previously described [16]. Significance level of each selected probes were shown in Supplementary Figure S 1. (B) MMCT of chromosome 14 was previously performed using HONE1 as the recipient cell line [18]. qPCR analysis of tumor-suppressive microcell hybrids (MCHs) and their tumor segregant (TS) cell lines, which are no longer tumor-suppressive, showed that NID2 expression was down-regulated in all five TS cell lines, when compared to their respective MCHs. Asterisk (*) indicates samples with more than two-fold differences compared to its MCHs.