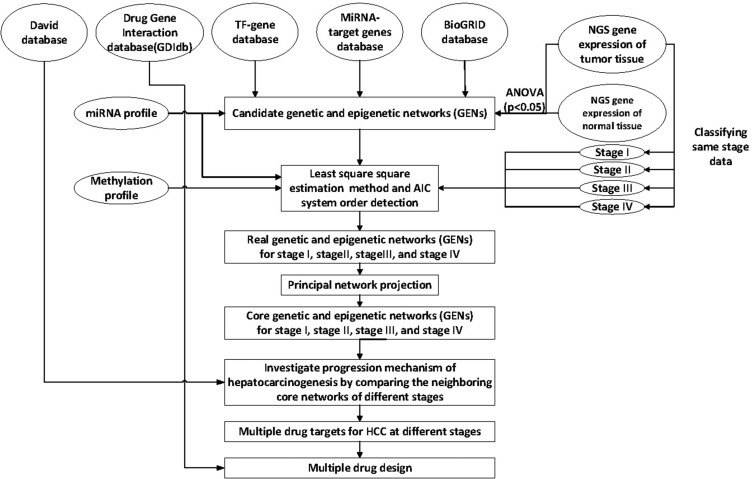
Figure 1. Overview of the construction of genetic and epigenetic networks (GENs) and core GEN to analyze the mechanism of hepatocarcinogenesis progression and multiple-drug design, via big database mining and system identification.
NGS profiles of HCC and normal tissue samples, miRNA and DNA methylation data, and the miRNA, TF-gene, and BioGRID databases were searched to construct candidate GENs, including candidate GRN, PPIN, and miRNA-gene regulatory networks. The false positive candidate GENs were pruned by system modeling and least square estimation, wherein the insignificant components were deleted from the system order based on AIC. The core GENs for each stage were obtained by extracting the principal structure of GENs through PNP. By mapping the core GENs to pathways retrieved using DAVID, and by comparing core GENs among the different stages, we investigated the mechanism of HCC progression. We also proposed potential therapeutic drugs against genes encoding significant epigenetic changes, in order to arrest the progression of HCC, by mining the DGIdb.