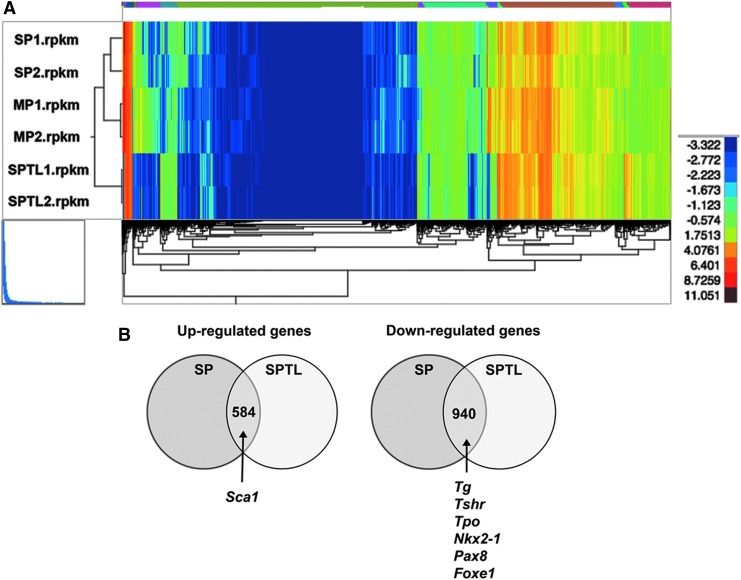
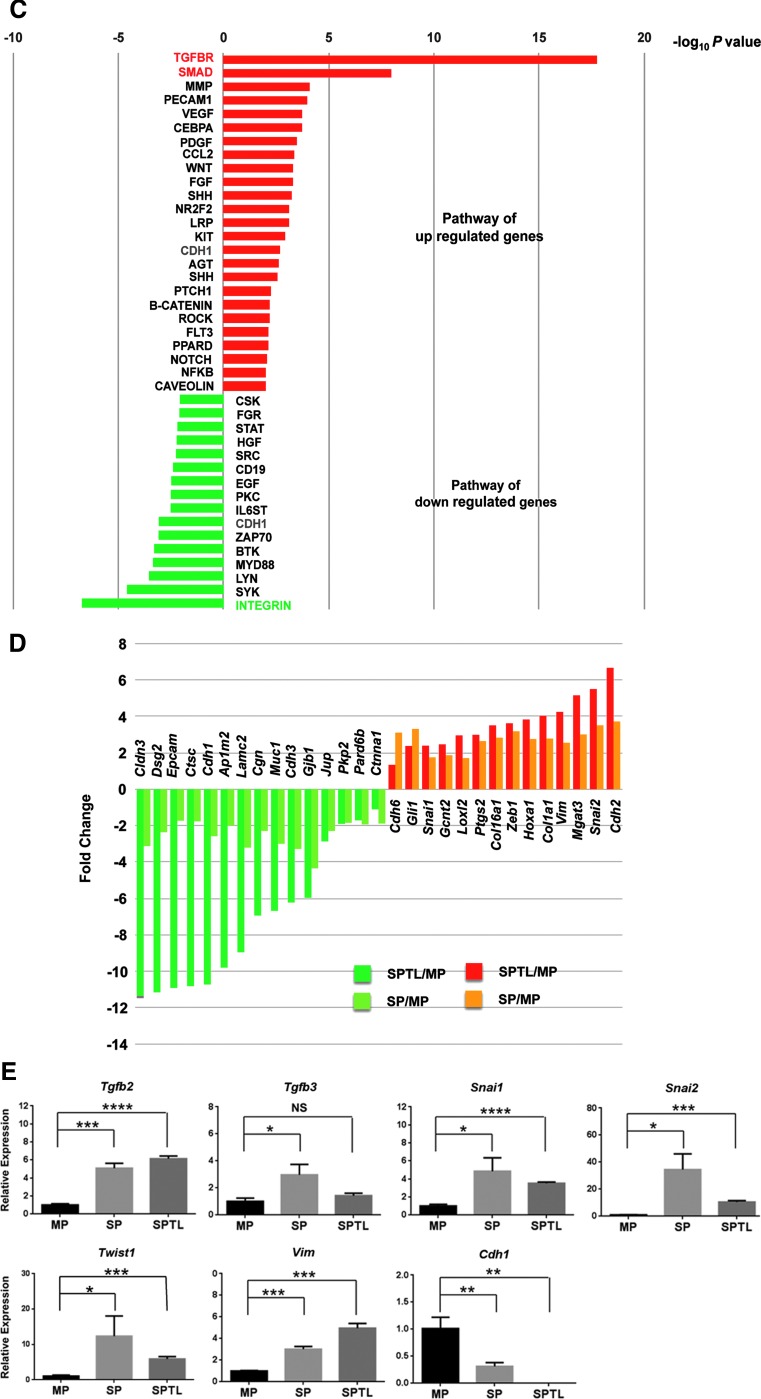
FIG. 5.
RNA seq analysis. (A) Dendrogram using total reads per kilobase million from SP1, SP2, MP1, MP2, and SPTL1, and SPTL2. (B) Venn diagram of commonly up- and downregulated genes between SP and SPTL cells compared with MP cells. Commonly upregulated genes include Sca1, while commonly downregulated genes include Tg, Tshr, Tpo, Nkx2-1, Pax8, and Foxe1 mRNAs. (C) Signal transduction pathways altered in SP and SPTL cells compared with MP cells were plotted based on –log10 p-values. Red bars are upregulated pathways, while green bars are downregulated pathways. The TGF-β and SMAD pathways (highlighted in red) are the two most upregulated pathways, while the integrin pathway (highlighted in green) is the most downregulated pathway. The cadherin pathway (CDH1, highlighted in blue) is both up- and downregulated. (D) Several genes in the cadherin pathways are plotted based on fold changes for SP/SPTL cells versus MP cells. (E) qRT-PCR analysis of several commonly up- and downregulated mRNAs using RNA isolated from MP, SP, and SPTL cells. The values obtained with the MP was set as 1. Mean ± SD in triplicate experiments are shown. Statistical significance between MP versus SP, or MP versus SPTL by unpaired t-test. *p < 0.05; **p < 0.005; ***p < 0.0005; ****p < 0.0001; NS, not significant.