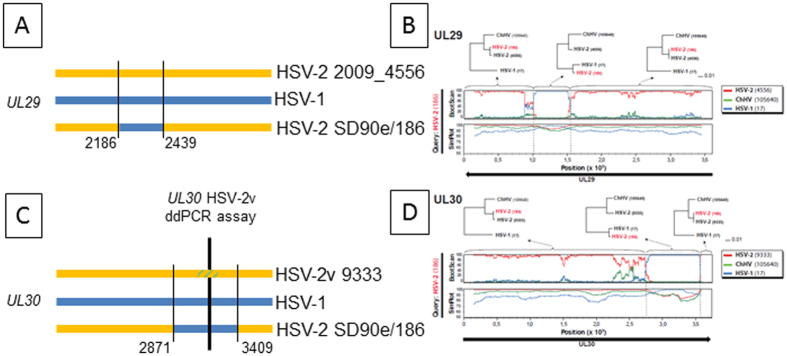
Figure 4. HSV UL29 and UL30 genotypes in strains circulating in humans.
(A and C) schematic diagrams of C-terminal coding sequences to approximate scale. HSV-1 is blue, rare HSV-2 strains are yellow. Blue bars in HSV-2 SD90e/186 represent circulating HSV-2 strains that have identity to HSV-1 in this region. Thin vertical black lines and associated strain 186 nucleotide numbers mark lateral flanks of HSV-1 identity. For UL30, thick vertical bar and yellow/green hatched zone within HSV-2v 9333 UL30 represent the locus detected by a ddPCR assay for which strain HSV-2v 9333 contains a variant nucleotide. (B,D) Recombination analysis of the UL29 and UL30 genes in HSV-2 strain 186. Bootscan and Simplot analyses are depicted for each gene. Clear shifts in bootstrap values supporting different phylogenetic topologies indicate recombination crossovers in both genes (also indicated by dotted lines). These crossovers are supported by the Simplot analysis, which demonstrates a shift in similarity with a higher similarity to HSV-1 in the recombination fragments. To further test and visualize ancestry of the recombination fragments, phylogenetic trees based on the recombination fragment and flanking regions are shown. HSV-2 strain 186 clearly shifts from clustering closely to HSV-2 in the trees based on the flanking regions, to clustering closely to HSV-1 in the trees based on the recombination fragments. These results suggest recombination with HSV-2 as major parental, and HSV-1 as minor parental strains.