Abstract
Development of gene therapy vectors requires cellular models reflecting the genetic background of a disease thus allowing for robust preclinical vector testing. For human p47phox-deficient chronic granulomatous disease (CGD) vector testing we generated a cellular model using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 to introduce a GT-dinucleotide deletion (ΔGT) mutation in p47phox encoding NCF1 gene in the human acute myeloid leukemia PLB-985 cell line. CGD is a group of hereditary immunodeficiencies characterized by impaired respiratory burst activity in phagocytes due to a defective phagocytic nicotinamide adenine dinucleotide phosphate (NADPH) oxidase. In Western countries autosomal-recessive p47phox-subunit deficiency represents the second largest CGD patient cohort with unique genetics, as the vast majority of p47phox CGD patients carries ΔGT deletion in exon two of the NCF1 gene. The established PLB-985 NCF1 ΔGT cell line reflects the most frequent form of p47phox-deficient CGD genetically and functionally. It can be differentiated to granulocytes efficiently, what creates an attractive alternative to currently used iPSC models for rapid testing of novel gene therapy approaches.
Chronic Granulomatous Disease (CGD) comprises a group of monogenetic immunodeficiencies characterized by impaired respiratory burst activity and microbicidal activity of phagocytes leading to recurrent life-threatening infections1. CGD can be cured by hematopoietic stem cell transplantation (HSCT)2. Alternatively, autologous retroviral gene therapy has been used in selected patients with X-linked CGD (X-CGD)3,4,5,6,7.
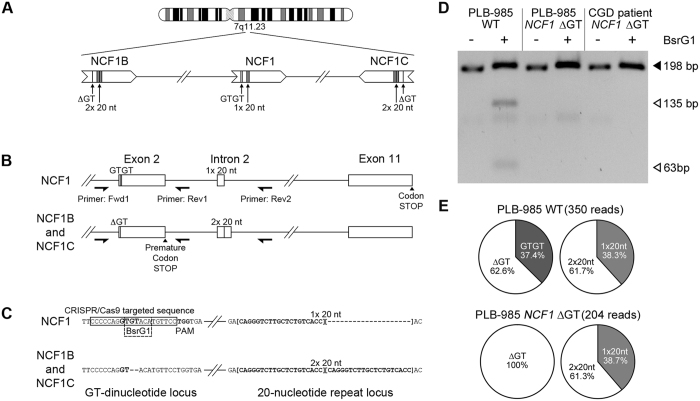
Mutations in gp91phox, p47phox, p67phox, p40phox or p22phox subunits of the phagocytic nicotinamide adenine dinucleotide phosphate (NADPH) oxidase may cause CGD8. In Western countries gp91phox-deficiency is the most frequent from (65%)9, followed by p47phox-deficiency (25%). Mutations within gp91phox are scattered throughout the whole cytochrome b-245 beta chain (CYBB) gene. Conversely, p47phox-deficiency is almost exclusively caused by a single GT-dinucleotide deletion (ΔGT) in exon 2 of neutrophil cytosolic factor 1 (NCF1) gene causing frameshift and premature translation stop. The ΔGT deletion is shared with two pseudogenes, NCF1B and NCF1C, all located on the same chromosome sharing extraordinary homology (99.5%) (Fig. 1A,B). Presumably, homologous recombination causes the predominance of the ΔGT mutation in p47phox-deficient CGD patients10.
Figure 1. NCF1 loci in PLB-985 cell lines.
(A) NCF1 gene is flanked by two pseudogenes, NCF1B and NCF1C. (B) Differentiators between NCF1 and pseudogenes. PCR primers: Fwd1, Rev1, Rev2. (C) DNA sequence of GT-dinucleotide and 20 nucleotide repeat loci. CRISPR/Cas9-targeted sequence, protospacer adjacent motif (PAM) and BsrG1 site: rectangles; 20-nucleotide repeat: squared brackets. (D) Loss of BsrG1 restriction site in PCR amplified NCF1 (B,C) exon 2 (primers Fwd1 and Rev1 (B)) in PLB-985 NCF1 ΔGT cells. Full-length gel is presented in Supplementary Fig. S6. (E) Percentages of reads from SMRT-seq of PCR products (primers Fwd1 and Rev2 (B)). The absolute number of analyzed SMRT sequencing reads which have passed all filtering criteria is given.
Currently, development of gene therapy vectors for p47phox-deficient CGD is hampered by the absence of human cell lines for rapid gene therapy vector testing. p47phox−/− mouse models exist, but cannot replace vector testing on human cells. Recently, we established human p47phox-deficient induced pluripotent stem cell (iPSC) lines harboring the ΔGT within the NCF1 gene11. Only these iPSC-based cell lines reflect the genetic background of the most frequent mutation in CGD. As maintenance and differentiation of iPSC lines is laborious and in many aspects impractical, we established a novel model for ΔGT p47phox-deficient CGD based on a human acute myeloid leukemia PLB-985 cell line12.
We nucleofected PLB-985 wild type (WT) cells with pPX458-NCF1 plasmid encoding Cas9, single guide RNA (sgRNA) targeting NCF1, and GFP. To introduce the ΔGT in NCF1 a 100-nucleotide long single stranded oligonucleotide (ssODN) template, carrying the ΔGT, was co-transfected (Fig. 1C).
Nucleofected GFP expressing cells were sorted by fluorescence-activated cell sorting (FACS) and expanded to monoclonal cell lines. Out of 609 sorted cells 22 clones could be expanded (3.6% survival rate, Supplementary Table S1). These clones were analyzed by PCR co-amplification of NCF1, NCF1B and NCF1C alleles (Fwd1, Rev1 primers, Fig. 1B), followed by BsrG1 digestion13. BsrG1 cleavage of NCF1 gene gives 135bp and 63bp products, while NCF1B, NCF1C, or mutated NCF1 lack the BsrG1 restriction site (Fig. 1C).
The BsrG1 analysis of one nucleofected PLB-985 clone displayed the same band pattern as of a ΔGT p47phox-deficient CGD patient (Fig. 1D and Supplementary Fig. S1), suggesting a Cas9-mediated disruption of the BsrG1 site in both NCF1 alleles (homozygous mutation efficiency 4.5%). The BsrG1 digestion analysis of the remaining clones suggested heterozygosity in these clones (Supplementary Fig. S1).
To confirm the presence of the ΔGT in mutated NCF1, genomic DNA of WT and pPX458-NCF1-treated PLB-985 cells were used for PCR co-amplification of the NCF1, NCF1B, and NCF1C (Fwd1, Rev2 primers, Fig. 1B). The barcoded PCR products were analyzed by single molecule real-time sequencing (SMRT-seq) for the presence of GT-dinucleotide, ΔGT, as well as for the presence of one copy of 20-nucleotide (20-nt) intronic repeat (1 × 20nt) derived from NCF1 or two repeats (2 × 20nt) derived from NCF1B or NCF1C (Fig. 1B). SMRT-seq analysis showed that PLB-985 WT and PLB-985 NCF1 ΔGT cell lines displayed almost identical percentage (61%) of reads with one or two copies of the 20-nucleotide repeat (Fig. 1E), indicating that all three NCF1 loci were co-amplified with comparable efficiencies. Moreover, all SMRT-seq reads from PLB-985 NCF1 ΔGT displayed the ΔGT, while in PLB-985 WT 37.4% harbored the GT-dinucleotide sequence, as expected from the two NCF1 alleles.
Potential off-target sites of utilized sgRNA were predicted (Supplementary Table S2) and DNA sequences of 14 sites with highest scores were analyzed for presence of insertions or deletions (indels) by Surveyor assay. In none of the potential off-target sites indels were observed (Supplemetary Fig. S2).
Flow cytometry analysis of granulocytically differentiated PLB-985 WT and PLB-985 X-CGD cells14 revealed 86.1% and 89.4% of p47phox protein expressing cells (Fig. 2A) (gating, Supplementary Fig. S3). No p47phox expression was observed in differentiated PLB-985 NCF1 ΔGT cells, which were CD14-negative, CD15-positive and partially gp91phox-positive (Supplementary Fig. S4).
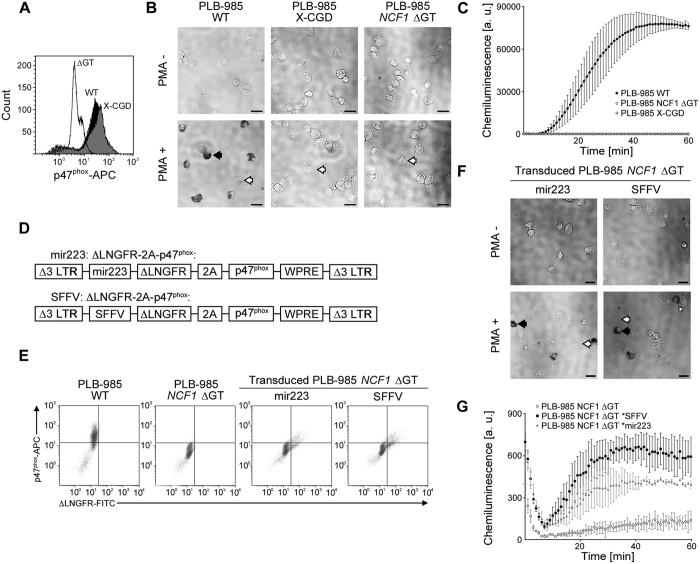
Figure 2. Superoxide production in PLB-985 NCF1 ΔGT.
(A) p47phox flow cytometry analysis and(B) ROS production in granulocytically differentiated PLB-985, PLB-985 X-CGD and PLB-985 NCF1 ΔGT cells measured by NBT test. NBT-positive: black arrow; NBT-negative: white arrows. Scale bar: 25 μm.(C) Chemiluminescence assay of un-transduced PLB-985 WT, PLB-985 NCF1 ΔGT, and PLB-985 X-CGD cell line (mean and standard deviation of mean, n = 3, a.u. = arbitrary units). (D) γ-retroviral vectors for reconstitution of p47phox expression. (E) Flow cytometry analysis of p47phox and ΔLNGFR expression in differentiated un-transduced PLB-985 WT and PLB-985 NCF1 ΔGT, and transduced PLB-985 NCF1 ΔGT cells. (F) ROS production in transduced PLB-985 NCF1 ΔGT cells upon differentiation, measured by NBT test. NBT-positive: black arrow; NBT-negative: white arrows. Scale bar: 25 μm. (G) Chemiluminescence assay of transduced PLB-985 NCF1 ΔGT cells (mean and standard deviation of mean, n = 3).
To confirm that differentiated PLB-985 NCF1 ΔGT cells mirror the absent respiratory burst observed in primary neutrophils of ΔGT p47phox-deficient CGD patients, we tested this cell line for NADPH oxidase-mediated superoxide production with nitroblue tetrazolium (NBT) test. In differentiated PLB-985 WT cells, phorbol-12-myristate-13-acetate (PMA)-stimulation induced superoxide production in 47.8 ± 4.2% of cells, as was visualized by formazan precipitates (Fig. 2B and Supplementary Fig. S5). In contrast, PMA-stimulation of differentiated PLB-985 X-CGD and PLB-985 NCF1 ΔGT cells resulted only in background signal of 1.7 ± 1.0% and 0.9 ± 0.5%. These results indicate that the established p47phox-deficient PLB-985 NCF1 ΔGT cell line does not respond to PMA-stimulation by reactive oxygen species (ROS) production. The absence of ROS production in PLB-985 X-CGD and PLB-985 NCF1 ΔGT cells was confirmed by chemiluminescence assay (Fig. 2C).
The iPSC model of ΔGT p47phox-deficient CGD11 can be differentiated to monocytes, macrophages11 and granulocytes15,16. Nevertheless, the use of iPSCs is laborious, and time-consuming, as phagocytic differentiation requires long culture periods of 35–43 days15,17 requiring large amounts of cytokines and continuous surveillance of the culture. In contrast full granulocytic differentiation of PLB-985 NCF1 ΔGT cell line takes 7 days, requires only fetal calf serum (FCS) restriction (5%) and supplementation with 0.5% N,N-dimethylformamide (DMF). Furthermore, the differentiation of neutrophils can be easily assessed by flow cytometry (Supplementary Fig. S3).
The function of the NADPH oxidase complex within PLB-985 NCF1 ΔGT cells was reconstituted by transduction with two γ-retroviral vectors encoding p47phox protein (Fig. 2D). Transgene expression was driven either by the ubiquitously active spleen focus-forming virus (SFFV) promoter or by the myelospecific microRNA 223 (mir223) promoter18. The expression cassette was composed of truncated Low-affinity nerve growth factor receptor (ΔLNGFR) and p47phox linked by the 2 A self-cleaving peptide19. Transduction efficiency reached 8.9% and 11.6% for mir223, and SFFV-driven γ-retroviral vectors (Fig. 2E).
Functional reconstruction of the NADPH oxidase complex activity in transduced and differentiated PLB-985 NCF1 ΔGT cells was assessed by NBT assay (Fig. 2F) and chemiluminescence assay (Fig. 2G). Percentage of NBT-positive cells reached 13.4 ± 2.1% with the mir223, and 11.8 ± 1.8% with the SFFV-driven ΔLNGFR-2A-p47phox expression constructs (Fig. 2F and Supplementary Fig. S5). Chemiluminescence assay of transduced and differentiated PLB-985 NCF1 ΔGT cells confirmed reconstitution of ROS production. These results clearly indicate that the defect of the NADPH oxidase complex caused by the ΔGT mutation within NCF1 in the PLB-985 NCF1 ΔGT cell line can be corrected.
In summary, the CRISPR/Cas9-generated PLB-985 NCF1 ΔGT cell line fully recapitulates the genetic background and functional NADPH oxidase defect found in majority of p47phox-deficient CGD patients, can be corrected genetically and differentiated into functional neutrophils. The PLB-985 NCF1 ΔGT cell line therefore represents a promising cost-effective tool for rapid gene therapy vector testing.
Materials and Methods
Reagents and antibodies
Single stranded DNA was purchased from Microsynth (Balgach, Switzerland). RPMI 1640 medium with stable glutamine and fetal calf serum (FCS) were purchased from PAN-Biotech (Aidenbach, Germany). Penicillin/streptomycin, Phusion High-Fidelity DNA Polymerase, and dNTPs were obtained from Thermo Fisher (Reinach, Switzerland), SCR7 was from Biovision (Milpitas, CA). BsrG1 was derived from New England Biolabs (Frankfurt/Main, Germany). N,N-dimethylformamide (DMF), propidium iodide (PI) and nitro blue tetrazolium (NBT) were obtained from Sigma Aldrich (Buchs, Switzerland). Mouse anti-human ΔLNGFR monoclonal antibody, clone ME20.4–1.H4, fluorescein isothiocyanate (FITC)-conjugated was purchased from Miltenyi Biotec (Bergisch Gladbach, Germany), and mouse anti-human p47phox monoclonal antibody clone 1 was allophycocyanin (APC)-conjugated by Becton Dickinson AG (Allschwil, Switzerland). PECy7-conjugated anti-mouse/human CD11b antibody was from BioLegend (Fell, Germany). Mouse anti-human CD15 FITC (Catalog no 347423) and mouse anti-human CD14 FITC (Catalog no. 345784) were from Beckton Dickinson AG (Allschwil, Switzerland). Mouse anti-Flavocytochrome b558 (human)-FITC clone 7D5 recognizing gp91phox was from LabForce AG (Muttenz, Switzerland).
Plasmid preparation
The sgRNA sequence targeting the NCF1 gene (but not NCF1B and NCF1C pseudogenes) was designed using Optimized CRISPR Design (http://crispr.mit.edu, F. Zhang laboratory, MIT 2015). Guide sequence CCCCCAGGTGTACATGTTCC was cloned into pSpCas9(BB)-2A-GFP (PX458) (F. Zhang, Addgene plasmid #48138)20 to generate pPX458-NCF1, which was confirmed by sequencing (Microsynth).
Cell culture and differentiation
We utilized PLB-985 cell line12, a subclone of HL-6021,22, as it is capable of granulocytic differentiation. Cells were cultured in RPMI 1640 medium supplemented with 10% (vol/vol) FCS, and 1% (vol/vol) penicillin/streptomycin in a humidified incubator at 37 °C and 5% CO2. Granulocytic differentiation of logarithmically growing PLB-985 cells at density of 0.8·106 cells/mL was induced by reduction of the FCS content to 5%, and supplementation with 0.5% (vol/vol) DMF. After 3 days, an equivalent of initial volume of differentiating medium was added and the differentiation continued until day 7.
CRISPR/Cas9 mediated generation of PLB-985 NCF1 ΔGT cell line
2·106 PLB-985 wild type (WT) cells were nucleofected (Amaxa Cell Nucleofector Kit V and Amaxa Nucleofector II, program C-023 (Lonza, Basel, Switzerland)) with 40 μg of the pPX458-NCF1 and a 100-nucleotide long ssODN sequence at final concentration 3 μM. The sequence of the ssODN was: GCC TCT TTG GAG GCT GAA TGG GGT CCC CCG ACT CTG GCT TTC CCC CAG GTA CAT GTT CCT GGT GAA ATG GCA GGA CCT GTC GGA GAA GGT GGT CTA CCG G. Immediately after nucleofection 500 μL of medium was added to the cuvette and the cells were incubated at room temperature for 10 minutes, transferred to 10 mL medium, then cultured for 48 hours. Additionally, the culture was supplemented with 1 μM SCR7 starting 3–4 hours post nucleofection. After 48 hours GFP expressing cells were sorted with FACS Aria III FCF (Becton Dickinson AG, Allschwil, Switzerland) into single wells with 100 μL of pre-conditioned, sterile filtered medium supplemented with 1 μM SCR7.
DNA Amplification and Restriction Fragment Length Polymorphism Analysis
Processing of human samples is covered by ethical vote KEK ZH nr. 2015/0135. Genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Hombrechtikon, Switzerland). Parallel PCR co-amplification of the NCF1, NCF1B, and NCF1C was performed using published primers23 (Fwd1 and Rev1, Fig. 1B). For PCR reaction using Fwd1/Rev2 primers Fwd1 primer was barcoded for each template producing products of 417 bp from NCF1 and of 435 bp from NCF1B and NCF1C. PCR products were pooled for SMRT sequencing and analyzed according to barcode identities.
The PCR mixture included HF 10× buffer, 200 μM of each dNTP, 240 nM of each primer, 0.04 U/μL of Phusion High-Fidelity DNA Polymerase, and 2.5 ng/μL of genomic DNA. Initial denaturation at 95 °C for 3 minutes was followed by 36 cycles of denaturation (95 °C, 30 seconds), annealing (70 °C, 30 seconds), elongation (72 °C, 8 seconds), and a final elongation step (72 °C, 1 minute). PCR products were digested with BsrG1 and visualized on a 3% (w/vol) agarose gel.
Single molecule real-time (SMRT) sequencing
The NCF1 gene and NCF1B and NCF1C pseudogenes were PCR amplified in parallel utilizing primers Fwd1 and Rev2 (Fig. 1B)23. The primer Fwd1 was barcoded for each FACS sorted clone and obtained PCR products were of 417 bp for NCF1 and 435 bp for NCF1B and NCF1C templates. PCR reaction consisted of initial denaturation (95 °C, 3 minutes), 40 cycles of denaturation (95 °C, 30 seconds), annealing (60 °C, 30 seconds), elongation (72 °C, 30 seconds), and a final elongation step (72 °C, 3 minutes). PCR products were gel purified using QIAquick Gel Purification Kit (Qiagen). 10–20 ng of gel-purified PCR products were pooled and analyzed by SMRT sequencing by Functional Genomics Center Zurich, ETH/University of Zurich, Zurich, Switzerland. Briefly, DNA amplicon librabry was produced using DNA Template Prep Kit 1.0 (Pacific Biosciences, Menlo Park, California, United States). The input DNA concentration and quality was measuered using Qubit Fluorometer dsDNA Broad Range assay (Life Technologies, Zug, Switzerland) and Bioanalyzer 2100 12 K DNA Chip assay (Agilent Technologies AG, Basel, Switzerland). The SMRT bell template was prepared by end-repair of the DNA amplicons, followed by blunt-end ligation of overhang adapters and exonuclease treatment. The SMRT bell template was complexed with polymerase using P6 DNA/Polymerase Binding Kit 2.0 (Pacific Biosciences) according to the manufacturer’s instructions. The samples were sequenced using Pacific Biosciences RS2 platform. From the raw reads, high quality circular consensus reads (CCS) were obtained through the Reads Of Insert protocol available in the SMRT Analysis suite (Pacific Biosystems). CCS reads were then de-multiplexed by exact matching of the sample barcodes, starting at the base immediately preceeding the Fwd1 primer sequence. Reads with a length between 400 and 450 nucleotides were retained and matched against the NCF1 reference sequence using blast24.
Surveyor Assay
Off-target sites were predicted utilizing the Optimized CRISPR Design (http://crispr.mit.edu, F. Zhang laboratory, MIT 2015). Loci of predicted sites (see Supplementary Table S2) were PCR amplified from gDNA of PLB-985 WT and PLB-985 NCF1 ΔGT cells. Corresponding PCR amplification products were mixed in a ratio of 1:1, while PCR amplification product of PLB-985 WT was used as a control. The samples were denatured at 95 °C for 10 minutes, slowly renatured, and digested using Surveyor® Mutation Detection Kit For Standard Gel Electrophoresis (Integrated DNA Technologies, Leuven, Belgium) according to the manufacturer’s instructions.
Data Availability
All SMRT sequencing data can be obtained upon request.
Virus production and transduction
Viruses were produced as described previously19. In brief, 293T cells were co-transfected with pUMVC, pMD2.VSV.G and γSIN-SFFV-ΔLNGFR-2A-p47phox 19 or γSIN-miR223-ΔLNGFR-2A-p47phox 18 in presence of 10 mM chloroquine. Sterile filtered virus containing supernatants were concentrated using Amicon-15 centrifugal filter devices with 100 kDa cutoff and stored at −80 °C until use.
PLB-985 NCF1 ΔGT cells were γ-retrovirally transduced with vectors co-encoding p47phox protein and low-affinity nerve growth factor receptor (ΔLNGFR)18,19 at concentration of 0.8·106 cells/mL, at multiplicity of infection (MOI) = 5, and in presence of 8 μg/mL protamine sulfate by spinoculation at 1286 g, 32 °C for 90 minutes, followed by incubation at 37 °C and 5% CO2 for 24 hours. Then, cells were differentiated into granulocytes in presence of 0.5% DMF and 5% FCS for 6 days and analyzed thereafter.
Flow cytometry analysis
ΔLNGFR surface staining was carried out with 6 μL of mouse anti-LNGFR-FITC (Miltenyi Biotec) per 5 * 105 cells in 100 μL for 20 minutes, followed by washing in PBS. Intracellular staining of p47phox was conducted utilizing 360 ng of mouse anti-p47phox-APC (clone 1) per 5 * 105 cells in 100 μL and the IntraCell Kit (Immunostep, Salamanca, Spain) according to the manufacturer’s instructions. Flow cytometry analysis was performed using a Gallios Flow Cytometer (Beckman Coulter).
Nitroblue-tetrazolium (NBT) test
Differentiated PLB-985 WT, PLB-985 X-CGD, and PLB-985 NCF1 ΔGT cells, as well as transduced PLB-985 NCF1 ΔGT cells were incubated in growth medium supplemented with 100 μg/mL NBT in the presence of 200 ng/mL PMA at 37 °C and 5% CO2 for 30 minutes. Subsequently, the cells were fixed in 1% (vol/vol) formaldehyde. 250 cells per cytospin slide were analyzed manually for NBT activity using a Leica DM IL Fluo light microscope equipped with a DFC420 digital camera and LEICA application suite acquisition software (Leica Microsystems AG, Glattbrugg, Switzerland).
Chemiluminescence assay
Chemiluminescence assay on differentiated PLB-985 WT, PLB-985 X-CGD, and PLB-985 NCF1 ΔGT cells, as well as transduced PLB-985 NCF1 ΔGT was conducted in 96-well plate format at a cell density of 1 * 105 cells/200 ml in a Mithras LB 940 Luminometer (Berthold Technologies GmbH, Zug, Switzerland) as described recently11.
Additional Information
How to cite this article: Wrona, D. et al. CRISPR/Cas9-generated p47phox-deficient cell line for Chronic Granulomatous Disease gene therapy vector development. Sci. Rep. 7, 44187; doi: 10.1038/srep44187 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the Functional Genomics Center Zurich for SMRT sequencing, and Giancarlo Russo for SMRT sequencing data analysis. The study was supported by The CGD Society (grant no. CGDS16/01), EU-FP7 CELL-PID HEALTH-2010-261387, EU-FP7 NET4CGD, Fondazione Ettore e Valeria Rossi foundation, Gebert Rüf Stiftung, program “Rare Diseases–New Approaches” (grant no. GRS-046/10), the Gottfried und Julia Bangerter-Rhyner-Stiftung, Hochspezialisierte Medizin Schwerpunkt Immunologie (HSM-2-Immunologie) and by ZIHP (Zurich Center for Integrative Human Physiology),
Footnotes
The authors declare no competing financial interests.
Author Contributions D.W. and U.S. performed experiments, analyzed results and made the figures; D.W., U.S. and J.R. conceived and designed experiments and wrote the manuscript.
References
- Seger R. A. Advances in the diagnosis and treatment of chronic granulomatous disease. Curr Opin Hematol 18, 36–41, doi: 10.1097/MOH.0b013e32834115e7 (2011). [DOI] [PubMed] [Google Scholar]
- Gungor T. et al. Reduced-intensity conditioning and HLA-matched haemopoietic stem-cell transplantation in patients with chronic granulomatous disease: a prospective multicentre study. Lancet 383, 436–448, doi: 10.1016/S0140-6736(13)62069-3 (2014). [DOI] [PubMed] [Google Scholar]
- Ott M. G. et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 12, 401–409, doi: 10.1038/nm1393 (2006). [DOI] [PubMed] [Google Scholar]
- Siler U. et al. Successful Combination of Sequential Gene Therapy and Rescue Allo-HSCT in Two Children with X-CGD - Importance of Timing. Curr Gene Ther 15, 416–427 (2015). [DOI] [PubMed] [Google Scholar]
- Kaufmann K. B. et al. Gene therapy for chronic granulomatous disease: current status and future perspectives. Curr Gene Ther 14, 447–460 (2014). [DOI] [PubMed] [Google Scholar]
- Bianchi M. et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood 114, 2619–2622, doi: 10.1182/blood-2009-05-221606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Niemiec M. J., Siler U., Urban C. F. & Reichenbach J. Restoration of anti-Aspergillus defense by neutrophil extracellular traps in human chronic granulomatous disease after gene therapy is calprotectin-dependent. J Allergy Clin Immunol 127, 1243–1252 e1247, doi: 10.1016/j.jaci.2011.01.021 (2011). [DOI] [PubMed] [Google Scholar]
- Curnutte J. T., Orkin S. H. & Dinauer M. C. In The molecular basis of blood disease 493–540 (Saunders, 1994). [Google Scholar]
- Roos D., van Bruggen R. & Meischl C. Oxidative killing of microbes by neutrophils. Microbes Infect 5, 1307–1315 (2003). [DOI] [PubMed] [Google Scholar]
- Gorlach A. et al. A p47-phox pseudogene carries the most common mutation causing p47-phox- deficient chronic granulomatous disease. J Clin Invest 100, 1907–1918, doi: 10.1172/JCI119721 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. et al. Derivation and functional analysis of patient-specific induced pluripotent stem cells as an in vitro model of chronic granulomatous disease. Stem Cells 30, 599–611, doi: 10.1002/stem.1053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. A., Lilly M. B., Heck L. Jr. & Rado T. A. Characterization of a new human diploid myeloid leukemia cell line (PLB-985) with granulocytic and monocytic differentiating capacity. Blood 70, 372–378 (1987). [PubMed] [Google Scholar]
- Vazquez N. et al. Mutational analysis of patients with p47-phox-deficient chronic granulomatous disease: The significance of recombination events between the p47-phox gene (NCF1) and its highly homologous pseudogenes. Exp Hematol 29, 234–243 (2001). [DOI] [PubMed] [Google Scholar]
- Zhen L. et al. Gene targeting of X chromosome-linked chronic granulomatous disease locus in a human myeloid leukemia cell line and rescue by expression of recombinant gp91phox. Proc Natl Acad Sci USA 90, 9832–9836 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann N. et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports 4, 282–296, doi: 10.1016/j.stemcr.2015.01.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney C. L. et al. Generation of functionally mature neutrophils from induced pluripotent stem cells. Methods Mol Biol 1124, 189–206, doi: 10.1007/978-1-62703-845-4_12 (2014). [DOI] [PubMed] [Google Scholar]
- Pedruzzi E., Fay M., Elbim C., Gaudry M. & Gougerot-Pocidalo M. A. Differentiation of PLB-985 myeloid cells into mature neutrophils, shown by degranulation of terminally differentiated compartments in response to N-formyl peptide and priming of superoxide anion production by granulocyte-macrophage colony-stimulating factor. Br J Haematol 117, 719–726 (2002). [DOI] [PubMed] [Google Scholar]
- Brendel C. et al. Human miR223 Promoter as a Novel Myelo-Specific Promoter for Chronic Granulomatous Disease Gene Therapy. Hum Gene Ther Methods 24, 151–159, doi: 10.1089/hgtb.2012.157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgensinger V., Seger R., Ryan M. D., Reichenbach J. & Siler U. Signed outside: a surface marker system for transgenic cytoplasmic proteins. Gene Ther, doi: 10.1038/gt.2010.73 (2010). [DOI] [PubMed] [Google Scholar]
- Ran F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308, doi: 10.1038/nprot.2013.143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. M. & Foster K. A. Differentiation of promyelocytic (HL-60) cells into mature granulocytes: mitochondrial-specific rhodamine 123 fluorescence. J Cell Biol 96, 94–99 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler H. G., Dirks W. G., Matsuo Y. & MacLeod R. A. F. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia 17, 416–426, doi: 10.1038/sj.leu.2402799 (2003). [DOI] [PubMed] [Google Scholar]
- Dekker J., de Boer M. & Roos D. Gene-scan method for the recognition of carriers and patients with p47(phox)-deficient autosomal recessive chronic granulomatous disease. Exp Hematol 29, 1319–1325 (2001). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SMRT sequencing data can be obtained upon request.