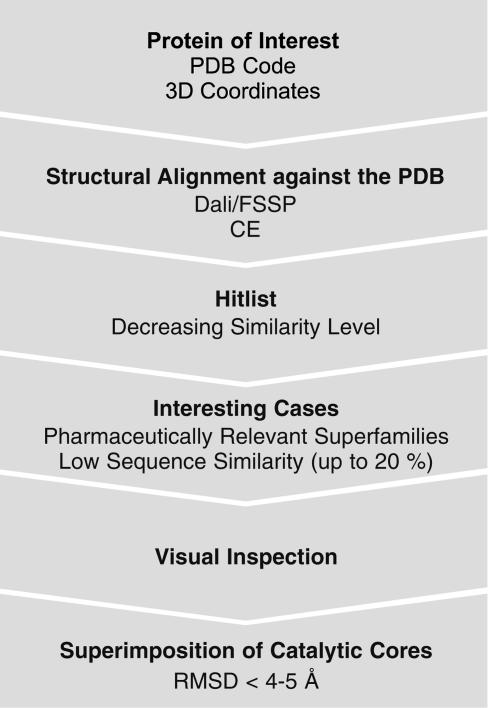
Fig. 1.
Database search strategy. Dali/Fold Classification Based on Structure–Structure Alignment of Proteins (FSSP) searches or Combinatorial Extension (CE) searches are performed by using the Protein Data Bank (PDB) code of the protein of interest. Alternatively, coordinates of a query protein structure may be submitted, and Dali/FSSP or CE compares them against those in the PDB. The FSSP and CE databases are based on exhaustive all-against-all 3D structure comparison of protein structures currently included in the PDB. Hits are listed with decreasing similarity level (3D and sequence similarity). From this list proteins belonging to pharmaceutically relevant families/superfamilies with low sequence identity (SI up to 20%) are chosen and visually inspected. The relevant part of the protein with respect to the delineated concept, i.e., the catalytic core, the conserved part of the domain where the active site is located, must be structurally similar and superimposed. RMSD, rms deviation for aligned Cα positions. When protein structures become too large most superimposition algorithms might fail so that smaller subsets of such big domains containing the interesting catalytic core have to be superimposed. Here, according to our experience, the CE algorithm (see Supporting Materials and Methods) delivers the best results.