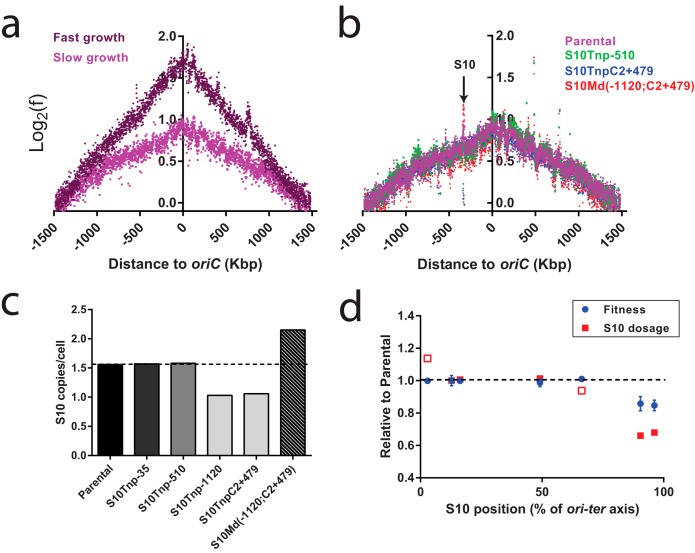
FIG 4 .
S10 relocation causes a dosage reduction in the absence of overlapping replication rounds that correlates with fitness impairment. MFA profiles are obtained by plotting the normalized number of reads at each position in the genome as a function of the relative position on the V. cholerae main chromosome with respect to ori1 (to reflect the bidirectional DNA replication) using an average of 1,000-bp windows. Windows containing repeat sequences were removed. (a) MFA profiles comparing the parental strain under fast- and slow-growth conditions, showing the presence (ori1 frequency, >2) and absence (ori1 frequency, <2) of multifork replication, respectively. (b) MFA profiles comparing the parental, movant, and merodiploid strains under slow-growth conditions. The arrow indicates the S10 position in the abscissa, evidencing its dosage alterations. (c) The S10 dosage for each strain in the absence of multifork replication was quantified with high precision using MFA data. The S10 dosage is the result of averaging the frequency of panels corresponding to the S10 locus and dividing it by the frequency of the 50 kbp flanking the ter1 zone. (d) The fitness and the S10 dosage of each movant in the absence of multifork replication were plotted as a function of the ori-ter axis of the V. cholerae genome. Blue circles correspond to the Wrel of each strain. Red squares correspond to the S10 dosage calculations from MFA experiments. Filled squares correspond to data directly obtained by MFA, while open squares mean that the data were inferred from the other data sets (see Materials and Methods).