Figure 2. DNA sequence validation, PCR identification of targeted SNVs and assessment of exon skpping.
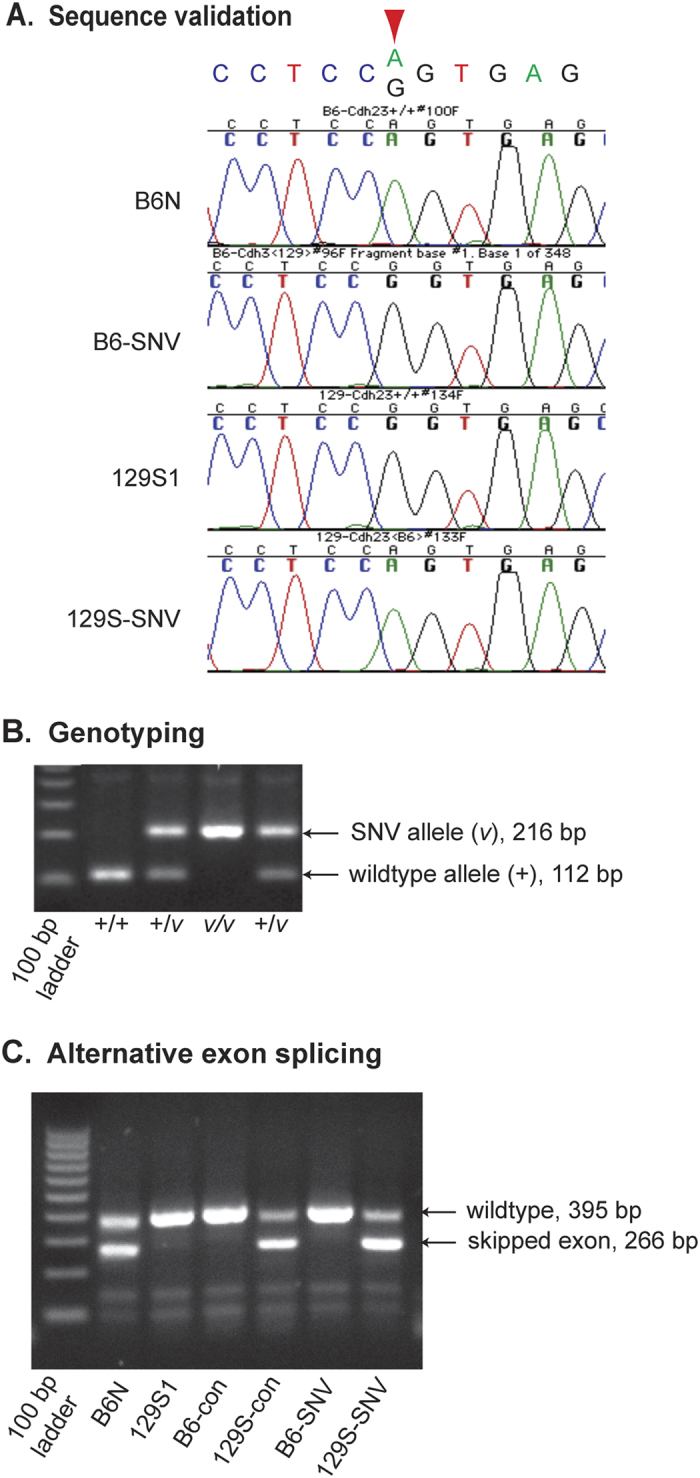
(A) Sequence chromatograms of PCR amplified DNA surrounding the targeted Cdh23c.753 nucleotide (indicated by the red downward-pointing arrow) confirm that C57BL/6 NJ (B6N) and 129S-Cdh23c.753A (129S-SNV) mice are homozygous for the Cdh23 c.753 A nucleotide, while 129S1/SvImJ (129S1) and B6N-Cdh23c.753G (B6N-SNV) mice are homozygous for the Cdh23 c.753 G nucleotide. (B) Identification of Cdh23 alleles with targeted SNVs by PCR amplification of the closely linked PGK-Neo insertion remnant. Primers flanking the PGK-Neo cassette insertion site were used to amplify PCR products that differ in size between the wildtype allele and the targeted SNV allele, which retains an intronic 104 bp remnant of the PGK-Neo cassette after Cre deletion. Because of its close proximity (178 bp) to the targeted SNV, the presence or absence of the PGK-Neo remnant can be used to distinguish the wildtype allele (+, 112 bp) from the targeted SNV allele (v, 216 bp). Lane 1, 100-bp DNA size ladder; lanes 2–5, genotypes of individual mice. (C) RT-PCR to evaluate the extent of exon skipping related to the Cdh23c.753A variant. cDNA primers flanking the alternatively spliced exon of Cdh23 (containing the c.753 nucleotide) were used to amplify alternatively spliced products. The PCR product size from the wild-type Cdh23 transcript (395 bp) is larger than the PCR product from the alternatively spliced, in-frame transcript (266 bp), which lacks the 129 bp skipped exon. Lane 1, 100-bp DNA size ladder; lane 2, B6N; lane 3, 129S1; lane 4, B6.129S1-Cdh23Ahl+ congenic (B6-con); lane 5, 129S1.B6-Cdh23ahl congenic (129S-con); lane 6, B6-Cdh23c.753G SNV (B6-SNV); and lane 7, 129S-Cdh23c.753ASNV (129S-SNV). Lanes 2, 5, and 7 show alternatively spliced transcripts caused by the Cdh23c.753A variant.
