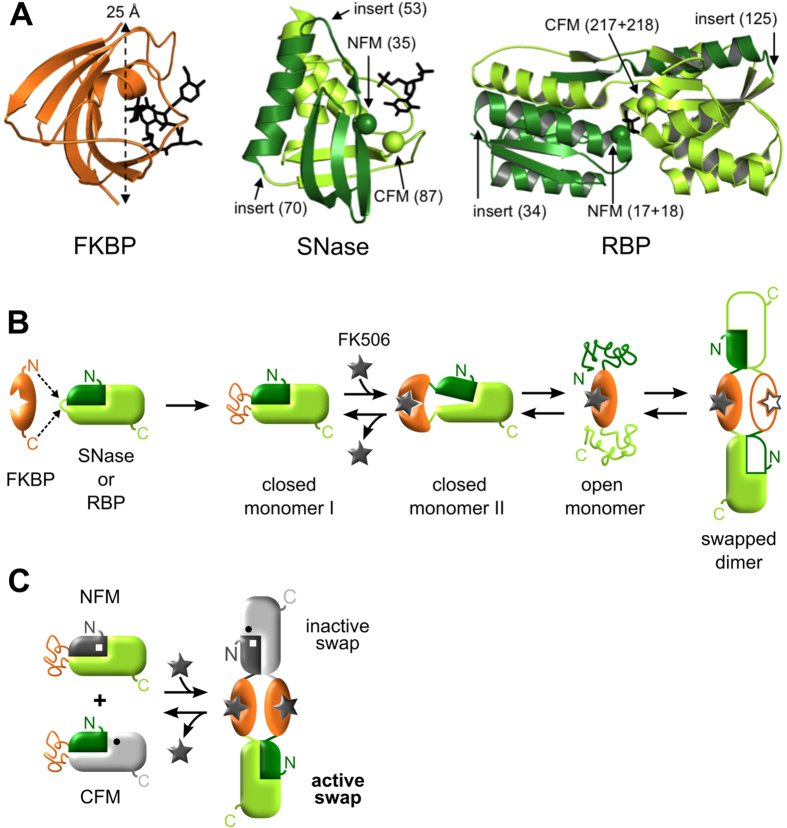
Figure 1. INDOS and domain-swapping bioswitches.
Lever proteins are colored orange and target proteins are colored green. (A) Structures of the lever protein FKBP (PDB 1FKF) with its N-to-C distance indicated, and the target proteins SNase (PDB 3BDC) and RBP (PDB 2IOY) with their insertion positions and functional mutations noted. Bound ligands (FK506, the inhibitor pdTp, and ribose) are shown as black sticks in the FKBP, SNase, and RBP structures, respectively. (B) Proposed mechanism of INDOS. A triggerable lever (FKBP) is inserted into a surface loop of a target protein (SNase or RBP). The target domain is initially more stable than the lever domain and the fusion protein adopts the closed monomer I conformation in the absence of FK506. FK506 binding stabilizes the lever, resulting in the strained and transiently populated closed monomer II state. Conformational strain is relieved by unfolding to the open monomer state followed by refolding in trans to generate swapped dimers and oligomers. (C) INDOS is used to create bioswitches by introducing NFM (white square) and CFM (black circle) point mutations into the target protein, on either side of the lever, rendering the construct inactive (partially or completely gray structures). The scheme in panel B results in swapped dimers, which can consist of NFM-CFM heterodimers (shown) and NFM-NFM and CFM-CFM homodimers (not shown). Biological activity is restored in NFM-CFM heterodimers (green domain), whereas all monomeric species as well as NFM-NFM and CFM-CFM homodimers remain nonfunctional.