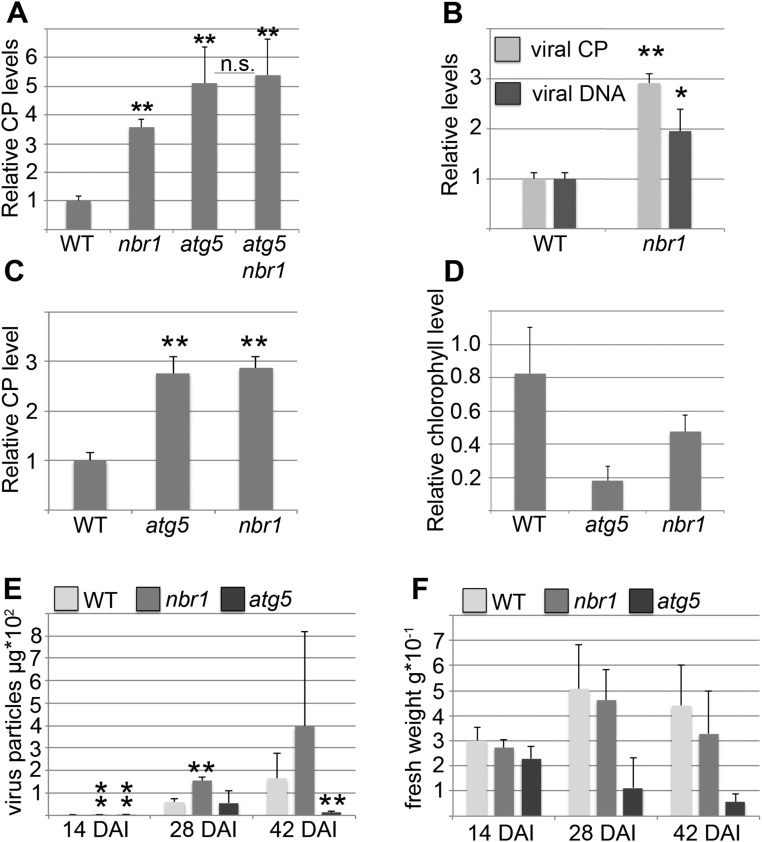
Fig. S1.
NBR1-dependent autophagy restricts CaMV infection in a persistent and isolate-independent manner, whereas bulk autophagy prevents disease-associated senescence and prolongs the timespan for virus production. (A) Capsid protein (CP) levels for CM1841 at 14 dai in nbr1 and atg5 single mutants and in atg5 nbr1 double mutants relative to WT plants. Values represent means ± SD (n = 4 biological replicates). (B) Viral titers of CaMV strain CM1841 in WT and nbr1 plants indicate that CaMV infection is suppressed similarly by NBR1-mediated autophagic processes at 28 dai and at 14 dai (Fig.1 C and D). Capsid protein and genomic DNA levels were determined by ELISA and qPCR, respectively; values represent means ± SD (n = 4) relative to WT plants. (C) Capsid protein levels of CaMV strain Cabb B-JI in WT, atg5, and nbr1 plants at 14 dai reveal that autophagy-mediated effects are not specific to the CM1841 strain (Fig. 1) or caused by the different inoculation methods (mechanical inoculation of purified Cabb B-JI particles vs. agroinfection of CM1841 DNA). Values are means ± SD (n = 4) relative to WT plants. (D) Ratio of total chlorophyll content in infected and noninfected plants. Error bars represent the SD (n = 6). (E) Capsid protein levels per individual CaMV Cabb B-JI–infected WT, atg5, and nbr1 plant at 14, 28, and 42 dai reveal that the total amount of virus accumulation is significantly reduced in atg5 plants at late stages of infection. Values represent means ± SD (n = 6) of virus particles per plant. (F) The fresh weight of plants was determined in parallel with the viral capsid protein levels in E, indicating premature death in atg5 plants. Values represent means of fresh weights for individual plants ± SD (n = 6). Asterisks indicate statistical significance (*P < 0.05; **P < 0.01) determined by Student’s t test (compared with WT); n.s., not significant.