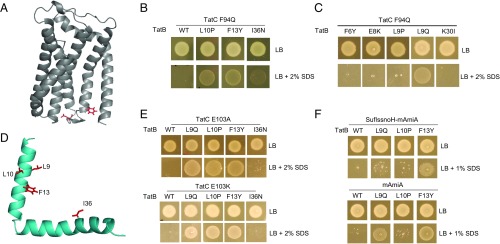
Fig. 2.
Isolation of suppressors of the TatC F94Q inactivating substitution. (A) Model of E. coli TatC (from ref. 47) showing the location of the F94 and E103 residues (in red) that form part of the signal peptide binding site. (B, C, and E) Growth of DADE (ΔtatABCD, ΔtatE) coproducing wild-type TatA alongside; (B and C) F94Q-substituted TatC or (E) E103A-substituted TatC, and the indicated substitution in TatB from plasmid pTAT101 on LB agar or LB agar containing 2% (wt/vol) SDS. (D) Structure of E. coli TatB (from ref. 68) with the locations of the TatCF94Q suppressor substitutions shown in red. (F) Strain MC4100 ΔamiA ΔamiC ΔtatABC coproducing the indicated TatB variants (with wild-type tatA and tatC) from pTAT101, alongside either a signal peptide variant of SufI lacking the h-region fused to the AmiA mature domain (SufIssnoH-AmiA), or the mature AmiA domain alone (mAmiA) on LB agar or LB agar containing 1% SDS. For all growth tests, a single colony of each strain/plasmid combination was resuspended in 30 μL of PBS and an 8 μL aliquot was spotted onto LB agar supplemented with appropriate antibiotics, along with SDS as indicated and incubated for 16 h at 37 °C.