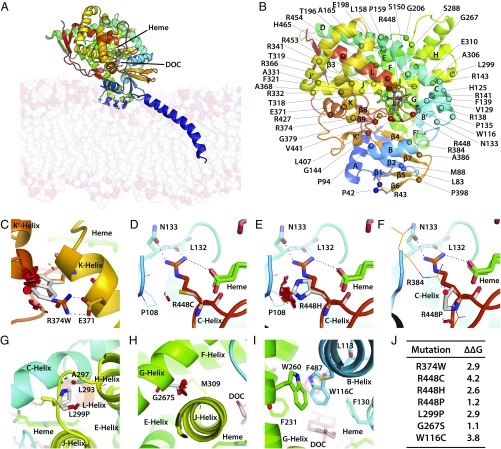
Fig. 2.
Modeling severe disruptive mutations affecting heme- or substrate-binding or CYP11B1 stability. Human CYP11B1 model constructed using the homologous CYP11B2 crystal structure as a template (PDB ID code 4DVQ), shown as side view showing the inner mitochondrial membrane bilayer and the bound heme (green) and deoxycorticosterone (DOC, pink) (A). Also shown are the positions of the α-helices, β-sheets, and the mutations analyzed in this study (B). R374 creates an ion-pair interaction with E371 at the end of the K-helix, which continues into a loop that forms a part of the heme-binding site (C). Mutation of this ion pair R374-E371 is disruptive. Mutations affecting the heme-binding site may not all be as disruptive, depending on the mutated residue. For example, residue R448, located on a loop between the meander region and L-helix, forms a part of the heme-binding site, and creates hydrogen bonds with the propionate tail of heme, the backbone atoms of L132, and the side chain of N133. Its mutation to Cys (D) or His (E) causes severe disruption, whereas substitution to Pro does not (F). Pro enhances rigidity of the loop, whereas the loop in the Cys/His mutants moves away from the vicinity of heme. L299 is located on the turn between the H- and J-helices, and its mutation to a more rigid Pro side chain decreases flexibility of the turn, thus impairing enzyme stability (G). G267S is located on the G-helix, and its mutation to Ser results in lost flexibility, as side chains of the bulkier Ser does not fit between G- and J-helices, causing steric clashes with residues on J-helix, significantly impairing enzyme stability (H). W116 is located on B′-helix, with its aromatic indole side chain surrounded by the aromatic side chains of F130, F231, W260, F287, and hydrophobic side chain of L113; these residues form the roof of the binding site of the aromatic substrate. A mutation to Cys disrupts the substrate-binding site (I). (J) The change (∆∆G kcal/mol) in protein stability upon mutation of each residue. A positive energy value indicates that the mutation is likely to be disruptive. The disks represent the pairwise atomic van der Waals radii overlap. Green disks represent almost in contact or slightly overlapping and red disks represent significant overlap.