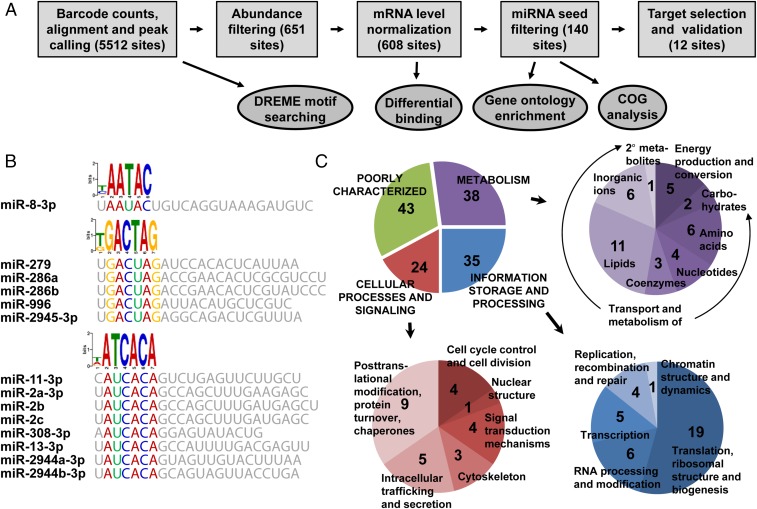
Fig. 3.
Identification of miRNA targets in the fat body by CLIP-seq. (A) Summary of the analysis pipeline. (B) Characteristic motif logos of 3′ UTR peaks. Characteristic reverse complement logos identified by DREME analysis of 3′ UTR peaks are shown with corresponding miRNA seed regions. Peak calling was performed from six AGO1 CLIP-seq libraries, and 3′ UTR peaks were used as input for DREME with a minimal core width as 6 nt. (C) Clusters of Orthologous Groups functional assignments for the 140 high-confidence targets. In the detailed illustration of cellular processes and signaling, two instances were each counted twice, because both were assigned to two different functional groups.