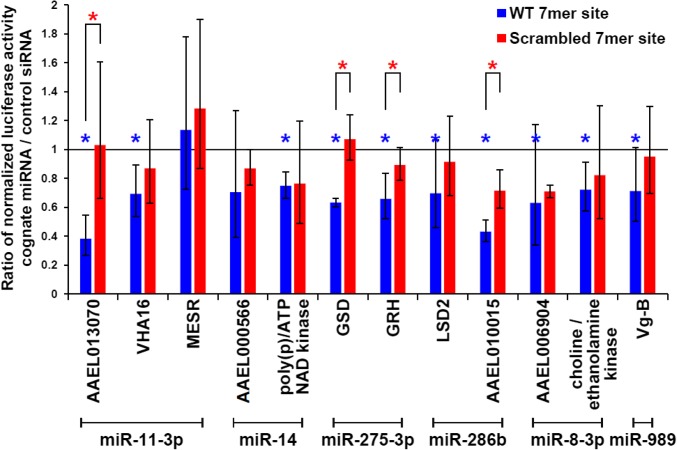
Fig. 4.
In vitro validation of miRNA targets. For each 3′ UTR, five and three biological replicates were measured for the wild-type (blue) and mutated (red) reporters, respectively. Statistical significance was calculated using unpaired one-tailed Student’s t tests. Data are represented as mean ± SD (*P value <0.05). Blue asterisks represent statistical significance in comparing the effect of miRNA mimic and negative control siRNA on wild-type reporters; red asterisks represent statistical significance between wild-type and mutated reporters with transfected miRNA mimic. Gene list is as follows: VHA16, AAEL000291, V-type proton ATPase 16-kDa proteolipid subunit; MESR, AAEL001659, misexpression suppressor of ras; poly(p)/ATP NAD kinase, AAEL000278; GSD, AAEL006834, glutamate semialdehyde dehydrogenase; GRH, AAEL000577, ortholog of D. melanogaster grainy head; LSD2, AAEL006820, lipid storage droplets surface binding protein 2; choline/ethanolamine kinase, AAEL008853; and Vg-B, AAEL006138, Vitellogenin isoform B.