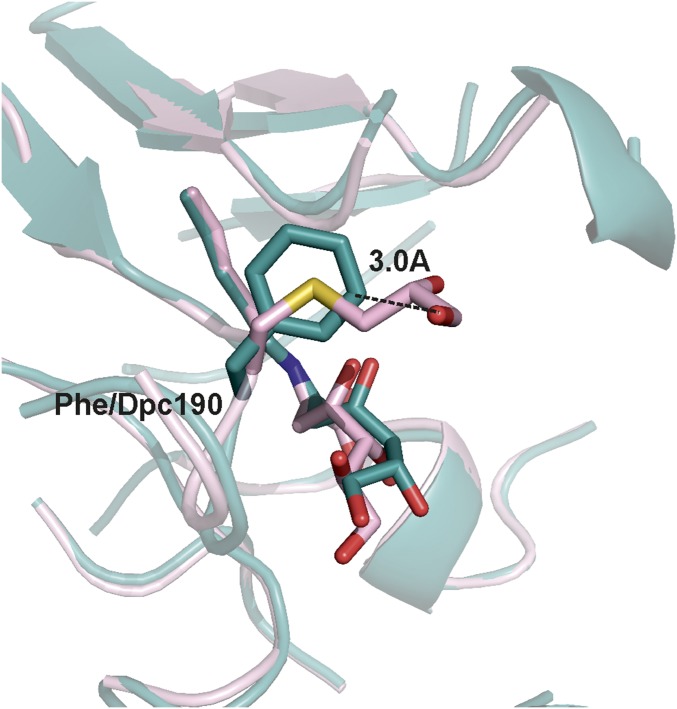
Fig. S7.
Comparison of the position of residue 190 in the Phe190Dpc and wild-type energy-minimized models. Computational energy minimization experiments were carried out using the Schrodinger Small Molecule Discovery Suite. The graphical user interface, Maestro (Schrödinger Release 2015-1: Maestro; Schrödinger, LLC), was used to model the Schiff bases formed between the product DHA and both the catalytic lysine of subunit B of the wild-type NAL and the catalytic lysine of subunit B of Phe190Dpc NAL. Energy minimization of the products formed was carried out using Macromodel with the OPLS_2005 force field (52) using water as the solvent. All side chains of amino acids within 6 Å of residues around the product were allowed to minimize, as well as the product itself.