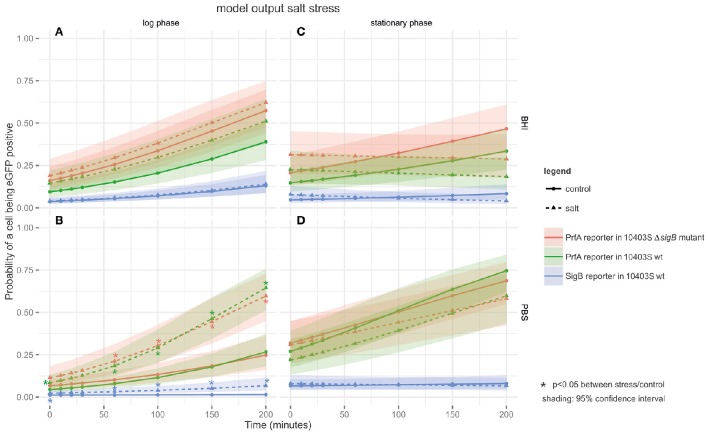
Figure 3.
Output of the linear model fitted to the salt stress FCM data. The panels are tiled to represent cultures grown to different growth phases in either BHI or PBS: (A) log phase cultures in BHI; (B) log phase cultures in PBS; (C) stationary phase cultures in BHI; and (D) stationary phase cultures in PBS. The y-axis represents the probability of a cell being eGFP-positive (as a result of active PrfA or σB), while the x-axis represents time in min. Three different strains are plotted on the graphs: the PrfA-reporter construct in a L. monocytogenes 10403S background, the PrfA-reporter construct in a L. monocytogenes 10403S ΔsigB mutant background, and the σB-reporter construct in a L. monocytogenes 10403S background. Dotted lines represent data collected under the stress condition (4.4% salt stress), the solid line represents the control condition (no salt), the shading around each line represents the 95% confidence interval. *Are color coded to match the strain they pertain to and indicate a significant difference in the probability that a cell is eGFP positive in the stress condition vs. the control condition within one strain at one time point.