Figure 2. FIGURE 2: In silico analysis of the CSP (EHI_016490) amino acid sequence.
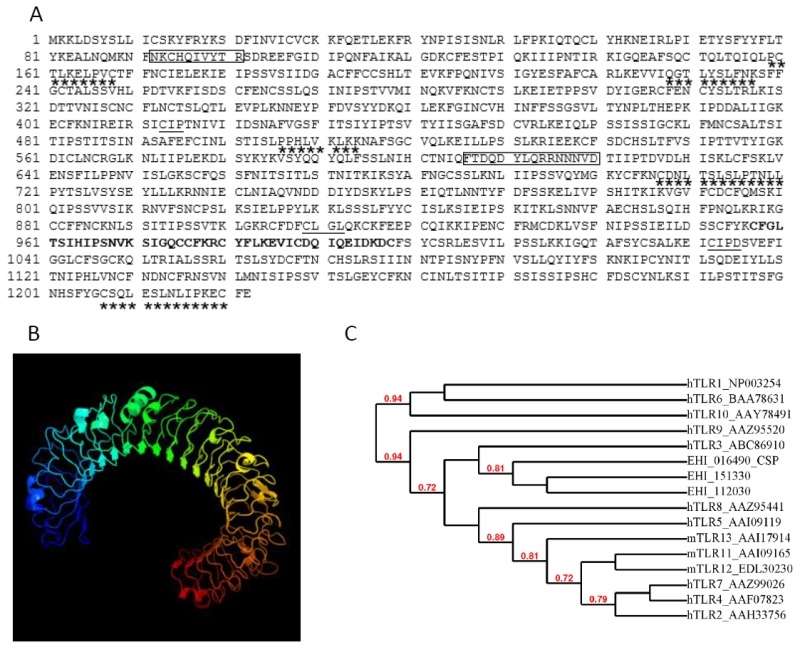
(A)Protein domains found in the amino acid sequence of EHI_016490 after scanning with InterProScan and SMART online tools. The TNFR_NGFR_1 family's cysteine-rich region signature is indicated in bold. The leucine-rich repeats (xxxLxxLxx) are underlined with stars, the prenylation sites (CaaX, where each “a” is an aliphatic amino acid) are underlined and the two peptides used to produce polyclonal rabbit serum are bordered. (B) A three-dimensional structure model of EHI_016490, generated using Phyre2. The model is based on the c1ziwA template (the structure of the human toll-like receptor 3 extracellular domain). In all, 856 residues (70% of the full sequence) were modelled with a confidence score of 99.9% for the highest-scoring template. (C) Dendogram showing the relatedness between EHI_016490, EHI_112030, EHI_151330 from Entamoeba histolytica , human TLR (hTLR) and murine TLR (mTLR). The number at each branch point represents percentage bootstrap support, calculated from 1000 replicates. Bootstrap values below 0.7 are not represented.
