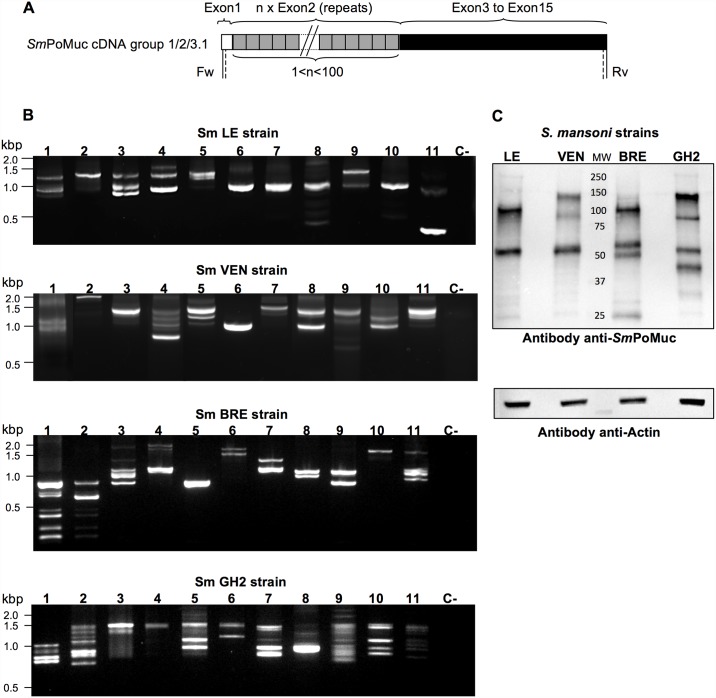
Fig 2. Diverse SmPoMucs are expressed by the different strains of S. mansoni.
(A) SmPoMuc cDNA structure. The 5’ region is characterized by a variable number of tandem repeats. Two different types of repeats (r1 and r2) were mainly identified in previous studies [19,20]. SmPoMuc cDNA structure is shared by previously published SmBRE, and SmGH2 strains [19,20] and the new ones, SmLE and SmVEN. The 3’ region (exon 3 to exon 15) harbors sequence differences that enable the proteins to be classified into the only three groups (groups 1, 2, and 3.1) known to be expressed [19,20]. (B) Transcription patterns of SmPoMucs in 11 individuals of each schistosome strain. Nested RT-PCR amplicons were obtained from individual sporocysts (1 to 11) of SmLE, SmVEN, SmBRE, and SmGH2, and resolved by agarose gel electrophoresis. PCR was performed using consensus primers that amplified the complete coding sequence of all SmPoMuc transcripts (located in exon 1 and exon 15; vertical and dashed lines represent the positions of the primers used for PCR and nested PCR, respectively). “C-” denotes the negative control. SmBRE and SmGH2 nested PCR results were from [21] (C) Inter-strain polymorphism at the protein level. Protein extracts (8 μg) were obtained from sporocysts of each S. mansoni strain, and Western blot analysis was performed using anti-SmPoMuc polyclonal antibodies. Actin was detected as a control.