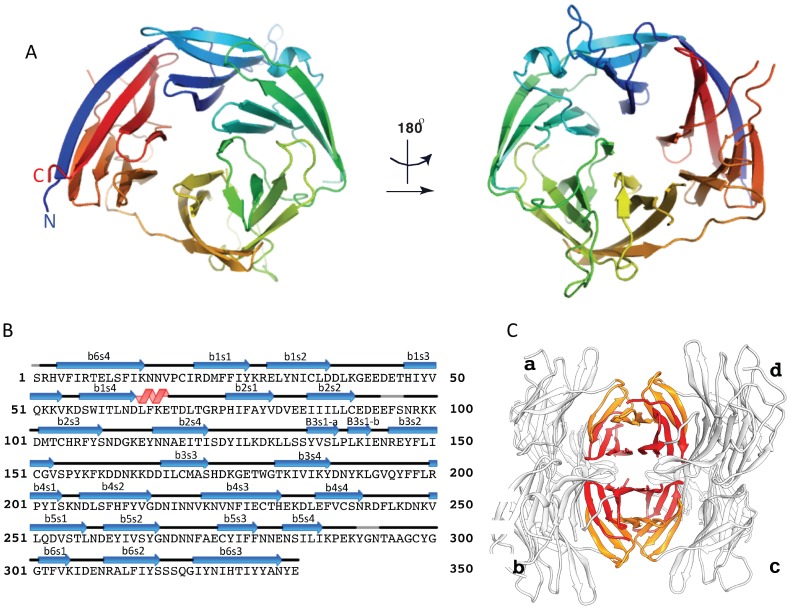
Figure 3. The crystal structure of the uncomplexed CyRPA.
(A) Orthogonal view of the ribbon representation of CyRPA, colored in rainbow fashion from the N-terminus (blue) through to the C- terminus (red). (B) Amino acid sequence and the secondary structure of CyRPA, showing the location of the 24 canonical strands of the six-bladed β-propeller [labelled ‘βmsn’ where the index n denotes the strand and the index m the sheet (n = 1,..,4; m = 1,..,6)]. (C) Assembly of the 222 pseudo-symmetric tetramer of CyRPA within the crystallographic asymmetric unit, showing the formation of the extended 5–5 and 6–6 sheets (orange and red, respectively). Each monomer of CyRPA in the asymmetric unit is labelled a, b, c, d.
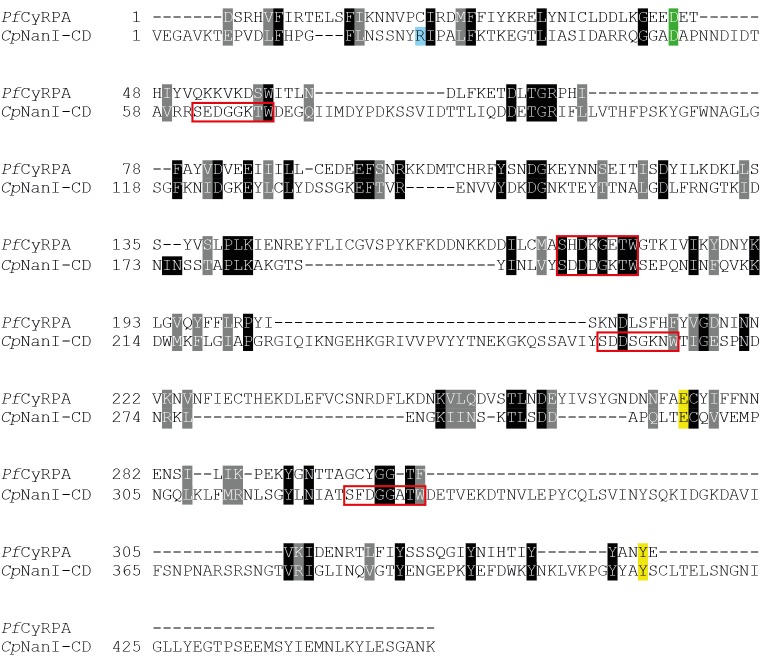
Figure 3—figure supplement 1. Comparison of CyRPA with C.
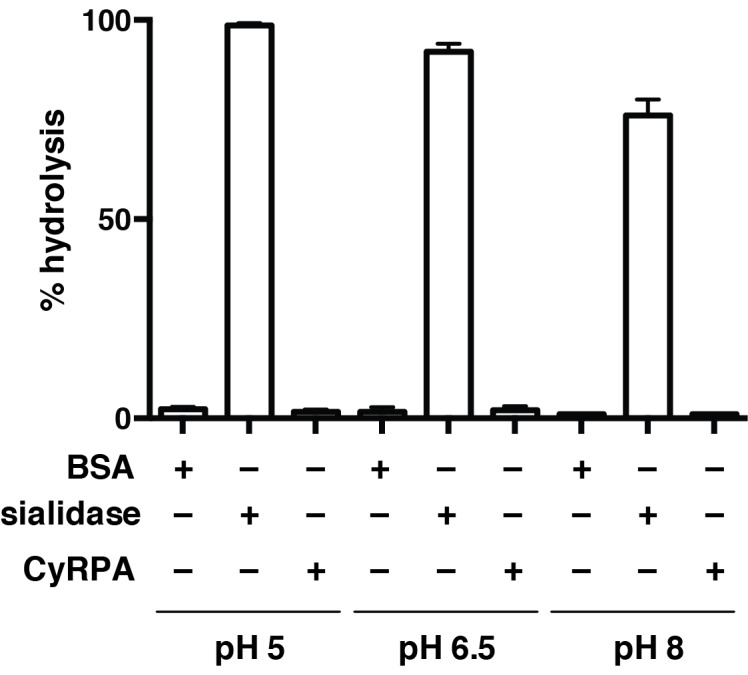
Figure 3—figure supplement 2. Recombinant CyRPA does not have sialidase activity.