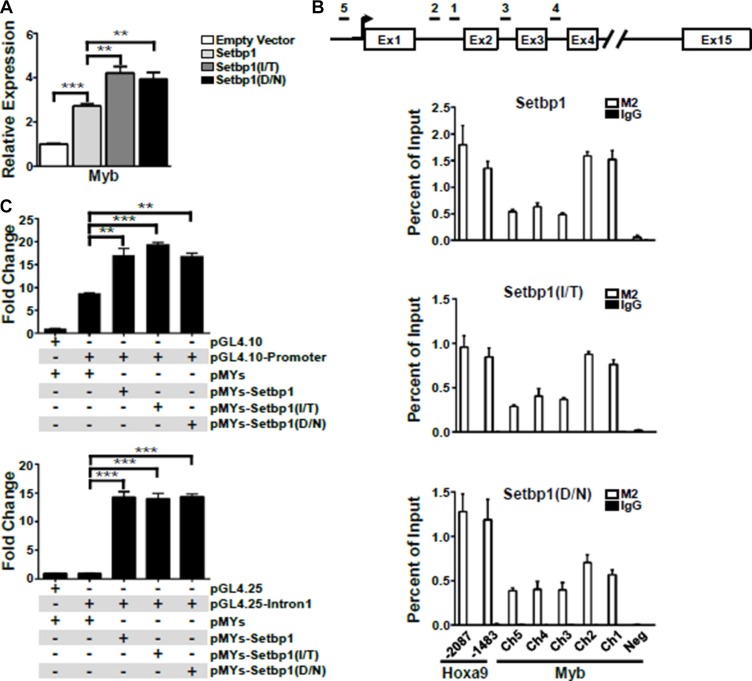
Figure 4. Myb is a direct transcriptional target of Setbp1 and Setbp1 missense mutants.
(A) Real-time RT-PCR analysis of Myb mRNA levels in mouse 5-FU treated BM progenitor cells purified 48 hrs after infection with the indicated retrovirus. Relative expression levels were calculated by normalizing to Gapdh mRNA levels in the same sample and also in cells infected by empty virus. The mean and SD of each relative expression level is shown. (B) Upper panel, schematic diagram showing locations of genomic regions used for chromatin immunoprecipitation (ChIP) analysis at the Myb locus. Lower panels, representative ChIP analyses of indicated regions of Myb locus (Ch1–5) and a negative control region (Neg, located at 10,179 bps upstream of Myb transcriptional start site) using anti-FLAG M2 antibody in 3xFLAG-tagged Setbp1 and Setbp1 mutants-immortalized cells followed by real-time PCR. (C) Dual luciferase assays of 293T cells transiently transfected with renilla luciferase reporter plasmid pRL-null, firefly luciferase reporter plasmids including pGL4.10 containing Myb immediate promoter (upper panel) and pGL4.25 containing the 1st intron of Myb (lower panel), along with the indicated pMYs construct. Mean and SD of ratios between firefly and renilla luciferase activity are shown. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t test).