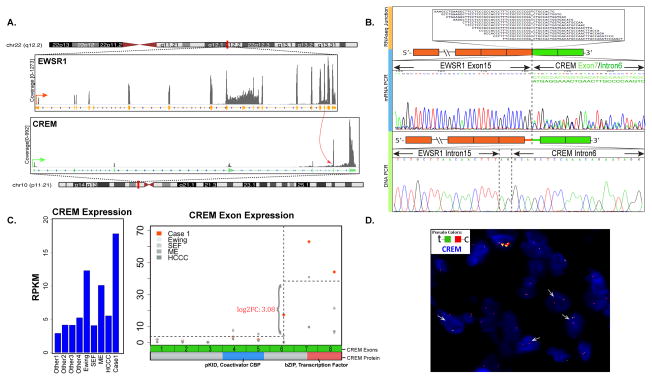
Figure 1. A novel EWSR1-CREM gene fusion was identified by transcriptome sequencing in index case 1.
(A) Fusion junction reads of paired-end RNAseq identified a candidate t(10;22) translocation resulting in an in-frame fusion between EWSR1 exon 15 and CREM exon 7. (B) This result was further confirmed by RT-PCR, with an alternative fusion transcript being identified secondary to alternative splicing. DNA breakpoints in EWSR1 intron 15 and CREM intron 6 was also identified by long range DNA PCR. (C) CREM exons 7 and 8, included in the fusion transcript, showed up-regulated expression compared to the CREM 5′ exons and other control samples with EWSR1 fusions (Ewing sarcoma, sclerosing epithelioid fibrosarcoma, myoepithelial tumor, hyalinizing clear cell carcinoma of salivary gland) or not (other 1–4). Among the control samples, one Ewing sarcoma with EWSR1-ERG fusion and one myoepithelial tumor with EWSR1-PBX3 fusion also showed elevated downstream CREM exons expression, possibly due to preferential expression of ICER isoform. CREM exons 7 and 8 encode the bZIP domain responsible for dimerization and DNA binding. pKID mediates interaction with the coactivator CBP. (D) Break-apart FISH assay showed CREM rearrangement with telomeric deletion.