Fig. 2.
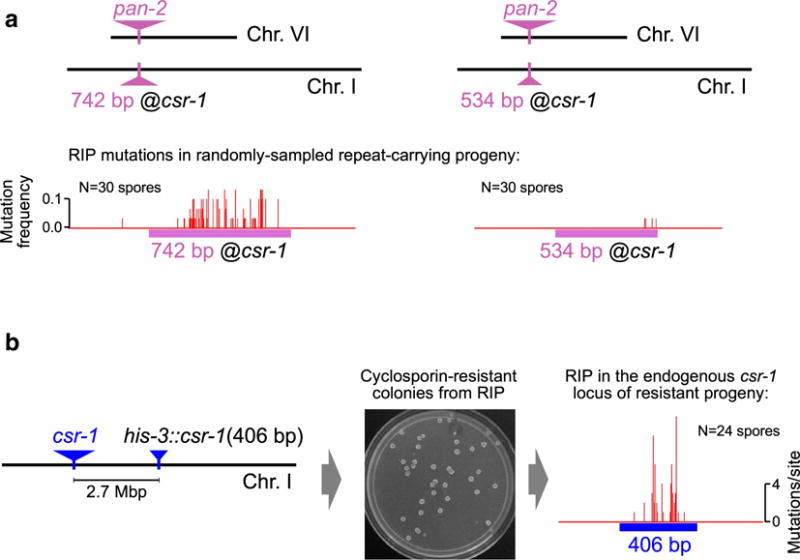
RIP detects widely separated repeats as short as 406 base-pairs. a Unlinked duplications of the pan-2 gene spanning 534 (right) and 742 (left) base-pairs are mutated by RIP. Fragments of the pan-2 gene corresponding to the chromosomal regions 3265392–3265925 (534 bp) and 3265184–3265925 (742 bp) on Supercontig_12.6 (Chr. VI) were cloned into the plasmid pEAG66 (Gladyshev and Kleckner 2014) between the restriction sites SacII and StuI and integrated into the standard strain FGSC#9720 (Colot et al. 2006) as the replacement of the csr-1 gene. Crosses were set up and mutation of the ectopic pan-2 copies was analyzed exactly as previously described (Gladyshev and Kleckner 2014). Thirty random “late-arising” progeny spores were examined in each case. The number of spores with at least one RIP mutation was 3 (10 %) for the 534-bp repeat, and 12 (40 %) for the 742-bp repeat. b 406-bp fragment of the csr-1 gene corresponding to the chromosomal region 7404971–7405376 on Supercontig_12.1 (Chr. I) was cloned into the plasmid pMF272 (Freitag et al. 2004) between the restriction sites NotI and EcoRI and integrated into the standard strain FGSC#9720 (Colot et al. 2006) near the his-3 gene, 2.7 Mbp away from the endogenous locus. Inactivation of the endogenous csr-1 gene by RIP produces cyclosporin-resistant progeny that can be selected by plating ejected spores en masse on sorbose agar with Cyclosporin A (5 μg/ml). 24 cyclosporin-resistant progeny were collected for analysis. Each sequenced csr-1 allele contained at least one mutation that could be attributed to RIP. No other sequence changes except those apparently produced by RIP could be detected, suggesting that mutation of the endogenous csr-1 gene can be used as a sensitive genetic test for RIP activity
