Fig. 3.
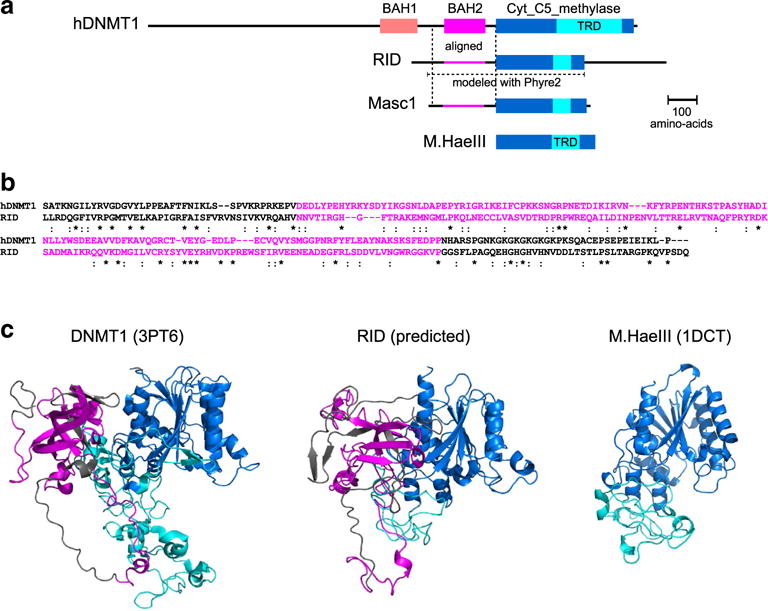
Neurospora RIP is mediated by a putative C5-cytosine methylase RID (RIP Deficient). a Structure of RID includes a conserved C5-cytosine methyltransferase domain (shown in blue) flanked by the N-terminal and C-terminal regions. The C-terminal region is absent in Masc1 (a homolog of RID that mediates a closely related phenomenon of “methylation induced premeiotically” in the fungus Ascobolus immersus). The difference in length between the methyltransferase domains of DNMT1, M.HaeIII and RID/Masc1 is largely explained by the reduction of the target recognition domain (TRD) in RID/Masc1 (shown in cyan). The structure of DNMT1 includes other conserved domains in the N-terminal extension that are omitted here for clarity. GenBank accession numbers are NP_001124295 (hDNMT1), XP_011392925 (RID), AAC49849 (Masc1), and P20589 (M.HaeIII). b N-terminal region of RID (also conserved in Masc1) can be fully aligned with a corresponding segment of the mammalian DNMT1 that includes a bromo-adjacent homology domain BAH2. This region was proposed to mediate putative interactions of DNMT1 with other proteins (Song et al. 2011) and double-stranded DNA (Song et al. 2012). c Predicted atomic structure of the conserved portion of RID (amino-acid positions 61-580): 97 % of residues are modeled at >90 % confidence with Phyre2 (Kelley et al. 2015). Representative structures of DNMT1 (PDB accession number 3PT6, amino-acid positions 928–1602) and M. HaeIII (PDB accession number 1DCT) are also provided. Domains are colored in accord with a and b
