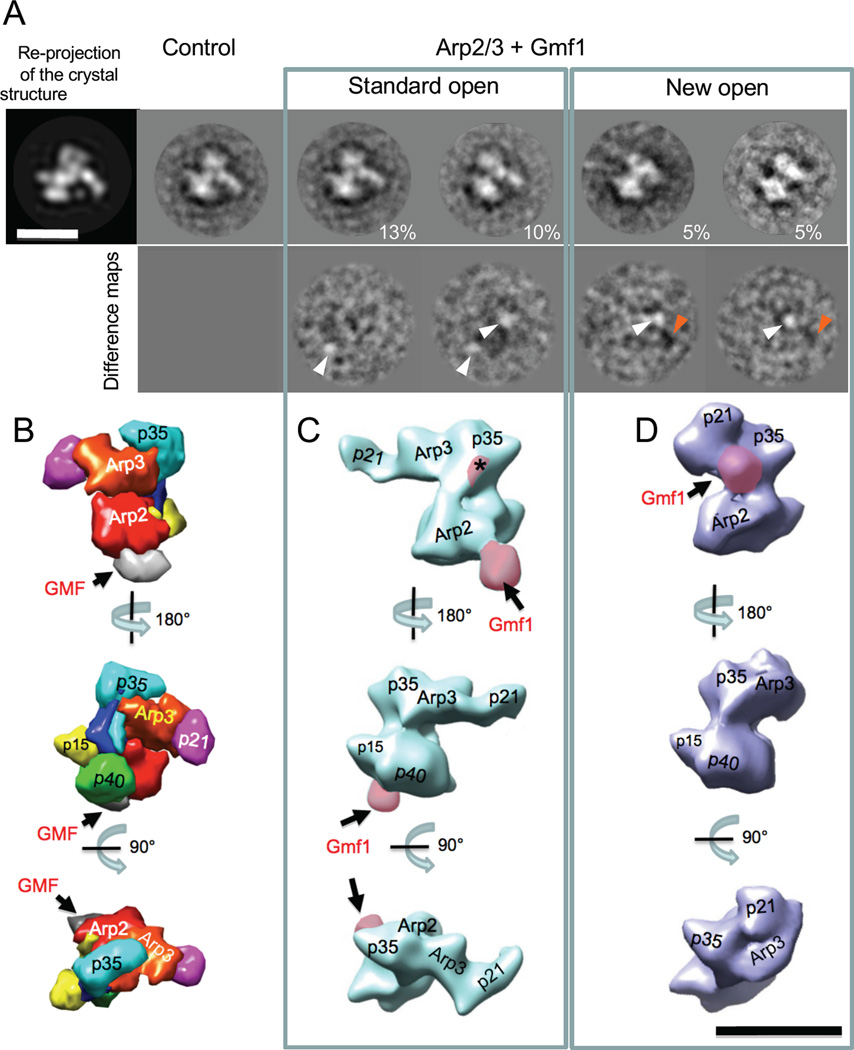
Figure 2. Three-dimensional structures of Gmf1-bound Arp2/3 complexes.
(A) Two-dimensional projections of Gmf1-bound Arp2/3 complex. Upper row, from left to the right: re-projection of the crystal structure of Arp2/3-GMF complex [14], free Arp2/3 2D projection in the same orientation, two representative 2D projections of Arp2/3-Gmf1 in standard open conformation, and two 2D projections of Arp2/3-Gmf1 in new open conformation. Numbers on the right reflect % of the particles in each class. Bottom row: difference maps, generated by subtracting the 2D projection of free Arp2/3 from Arp2/3-Gmf1 projections, respectively. White arrowheads highlight adding of the new mass, orange arrowheads highlight loss of the mass. (B) Crystal structure of GMF-bound Arp2/3 complex [14] filtered to 25 Å resolution using UCSF Chimera [51]. Arp2/3 complex subunits, color-coded and labeled as in Figure 1A. Arrows highlight the position of GMF (grey) bound to Arp2 and p40/ARPC1 subunits. (C,D) Three-dimensional reconstructions of Gmf1-bound Arp2/3 complex in the standard open conformation (C) and the new open conformation (D). Additional densities, approximately the size of Gmf1, are shaded in pink and labeled with arrows. Asterisk in (C) marks a new mass at the proposed second binding site for GMF on Arp3. Three separate views of Arp2/3 complex shown in B-D are aligned to each other for comparison. Scale bar, 10 nm.