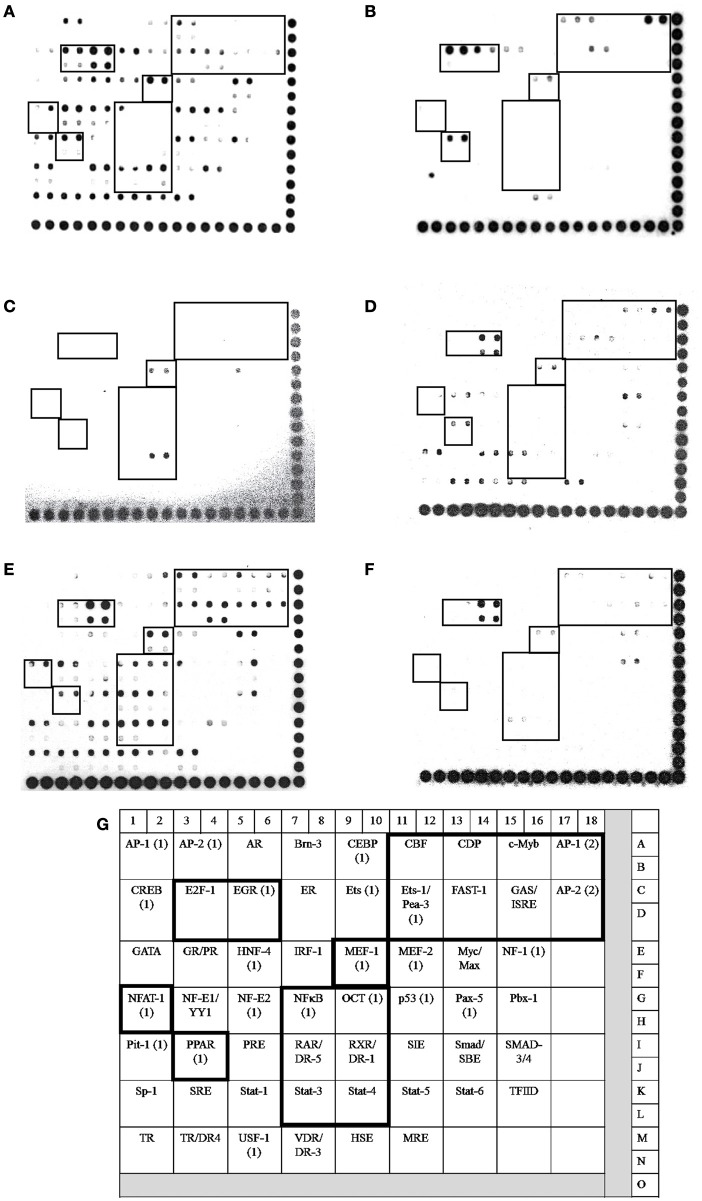
Figure 4.
Transcription factor activation by NaO in S. aureus-challenged bMECs. Protein/DNA array blots were used to analyze 56 different transcription factor DNA-binding sites from samples that were obtained from (A) bMEC nuclear extracts (control), (B) bMECs that were challenged with S. aureus, (C,E) bMECs that were treated with 0.25 or 1 mM NaO for 24 h, and (D,F) cells that were treated with 0.25 or 1 mM NaO and challenged with S. aureus for 2 h. (G) Schematic diagram of transignal™ protein/DNA array I. The DNA samples were spotted in duplicate in two rows (top: Undiluted; bottom: Dilution 1/10). Biotinylated DNA was spotted for alignment along the right and bottom sides of the array. TFs whose activation status was affected by NaO are highlighted with boxes. AP-1, activating-protein 1; AP-2, activating enhancer binding protein 2; CBF, core binding factor; CDP, CCAAT displacement protein; c-Myb, c-myeloblastosis transcription factor; DR, death receptor; EGR-1, early growth response protein 1; Ets-1, Ets-1 transcriptional factor; E2F-1, E2F transcription factor 1; FAST-1, forkhead activin signal transducer-1; GAS, interferon-gamma activated sequence; ISRE, interferon-stimulated response element; MEF-1, myeloid Elf-1 like factor; NF-κB, nuclear factor-κB; NFAT-1, nuclear factor or activated T cells; PPAR, peroxisome proliferator-activated receptor; OCT-1, octamer transcription factor; RAR, retinoic acid receptor; RXR, retinoid X receptor; Stat, signal transducer and activator of transcription.