Abstract
Gene products of the major histocompatibility complex (MHC) of human and non-human primates play a crucial role in adaptive immunity, and most of the relevant genes not only show a high degree of variability (polymorphism) but also copy number variation (CNV) is observed. Due to this diversity, MHC proteins influence the capability of individuals to cope with various pathogens. MHC and/or MHC-linked gene products such as odorant receptor genes are thought to influence mate choice and reproductive success. Therefore, MHC typing of wild and captive primate populations is considered to be useful in conservation biology, which is, however, often hampered by the need of invasive and time-consuming methods. All intact Mhc-DRB genes in primates appear to possess a complex and highly divergent microsatellite, DRB-STR. A panel of 154 pedigreed olive baboons (Papio anubis) was examined for their DRB content by DRB-STR analysis of genomic DNA. Using the same methodology on DNA of feces samples, DRB variability of a silvery gibbon population (Hylobates moloch) (N = 24), an endangered species, could successfully be studied. In both species, length determination of the DRB-STR resulted in the definition of unique genotyping patterns that appeared to be specific for a certain chromosome. Moreover, the different STR lengths were shown to segregate with the allelic variation of the respective gene. The results obtained expand data gained previously on DRB-STR typing in macaques, great apes, and humans and strengthen the conclusion that this protocol is applicable in molecular ecology, conservation biology, and colony management, especially of endangered primate species.
Electronic supplementary material
The online version of this article (doi:10.1007/s00251-016-0968-0) contains supplementary material, which is available to authorized users.
Keywords: Primates, MHC, Microsatellites, Conservation biology, Pre-clinical research
Introduction
The major histocompatibility complex (MHC) class I and II molecules are cell-surface glycoproteins, which play a central role in immune-related processes as in control or susceptibility to infectious diseases, in transplantation research, and in reproduction immunology by presenting peptides to CD8 and/or CD4 T lymphocytes. The MHC genes/loci encoding these molecules are structured in gene families, the hallmark of which is the allelic and copy number variation (CNV) of most of their genes. Balancing selection exerted mainly by pathogen pressure is believed to represent one of the major forces driving MHC polymorphism and diversity (Bernatchez and Landry 2003; Piertney and Oliver 2006; Spurgin and Richardson 2010). Characterization of polymorphic and polygenic MHC genes is therefore an excellent measurement of genetic health and as such of value in conservation biology, as e.g., management of captive breeding programs for endangered species (Cai et al. 2015; Hans et al. 2015; Oliver and Piertney 2012; Pechouskova et al. 2015). One of the most variable MHC genes in many vertebrate species is the DRB gene encoding the beta chain of the class II DR molecule. Therefore, DRB typing has often been used as an indication for MHC diversity in various species and for diverse purposes, as e.g., the importance of the MHC for mate choice (Setchell et al. 2013; Setchell et al. 2011), heterozygous advantage testing against heterogenous pathogen pressures (Oliver et al. 2009; Osborne et al. 2015), identification of selective pressures acting on MHC haplotypes (Huchard et al. 2008), pathogen-driven balancing selection (Nishita et al. 2015), selection countering drift to maintain MHC polymorphism (Oliver and Piertney 2012), and understanding genetic variability for the management of captive breeding programs (Cai et al. 2015).
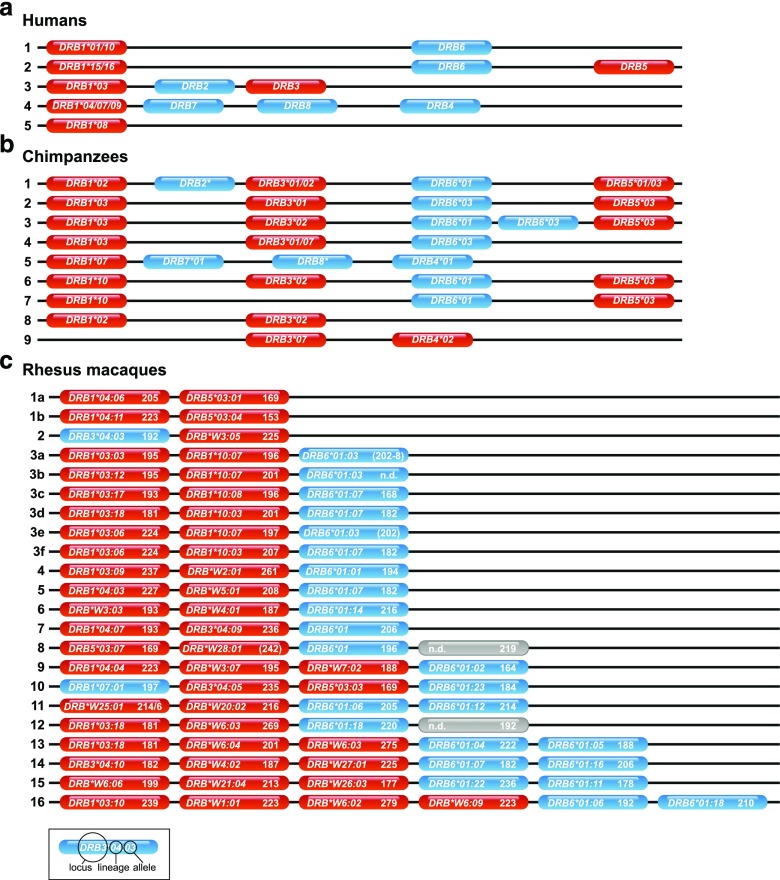
In primates, the DR region is composed of an utmost oligomorphic DRA gene encoding the alpha chain, and polymorphic DRB gene(s) encoding one or several beta chains. In humans, for example, five DR region configurations have been defined. Each configuration is composed of a DRA gene in conjunction with a unique combination of DRB genes/lineages, which are usually inherited together as a so-called haplotype. A lineage is defined as group of alleles (allelic variants) within a given gene that cluster together in the phylogenetic tree and are thus supposed to have the same evolutionary origin. The high degree of human DRB variability is mainly due to its polymorphic (variable) DRB1 gene. Chimpanzees, as representatives of the great apes and the closest relatives of humans, show a higher number of region configurations, only three of which displaying allelic variation. This allelic variation is, however, not unique to the DRB1 gene as polymorphism is present within all genes. In contrast, Old World monkeys (OWM) as macaques show a far higher number of region configurations with a very low degree of allelic variation. This peculiar situation of CNV in OWM is the reason, why DRB typing in these animals is hampered by the need of invasive DNA/RNA isolation procedures from blood samples as well as subsequent laborious and expensive techniques such as cloning/Sanger sequencing or next-generation sequencing. However, in close proximity to the highly variable DRB exon 2 is a complex microsatellite located in intron 2. This so-called DRB-STR (D6S2878) is present in all primate DRB genes analyzed so far with an intact exon 2–intron 2 boundary (Andersson et al. 1987; Bergstrom et al. 1999; Epplen et al. 1997; Riess et al. 1990; Trtkova et al. 1995). In humans, chimpanzees, and macaques, it was recently proven that the length variability of the DRB-STR is in accordance with the allelic variation of the DRB gene. Moreover, DRB typing with this specific microsatellite resulted in unique genotyping patterns that appear to be specific for a given DRB haplotype as illustrated for DRB haplotyping in Indian rhesus macaques (see Fig. 1c) (de Groot et al. 2008; Doxiadis et al. 2007; Doxiadis et al. 2013).
Fig. 1.
Schematic representation of DRB region configurations of a humans, b chimpanzees, and c Indian rhesus macaques. For humans and chimpanzees only, region configurations are presented, whereas for Indian rhesus macaques also, allelic variations are shown accompanied with the observed STR length. Blue = pseudogenes, red = genes, gray = allele not defined (n.d.)
Based on these published data, we examined the possibility to type for DRB by using the DRB-STR microsatellite in another Old World monkey species, the olive baboon. Very little was known so far about the DRB allelic variation and haplotype composition of the olive baboon (Papio anubis, Paan-DRB), although the olive baboon serves as model species in renal (Poirier et al. 2015), and xeno-transplantation research (Le Bas-Bernardet et al. 2015), and for immunotoxicity studies (Poirier et al. 2014).
An endangered lesser ape species, the silvery gibbon shares a common ancestor with humans ∼18–20 mill years ago (Perelman et al. 2011). The natural habitat of the silvery gibbons is the island of Java, where they occupy lowland and hill and lower mountain rainforests. A population survey of the silvery gibbon carried out between 1994 and 2002 found that 4000 to 4500 individuals were present not only in habitat fragments in the west of Java but also in central Java (Nijman 2004). The rate of which its numbers appear to be declining, mainly due to deforestation, the silvery gibbon has a high chance of going extinct. There are three wild populations located in Java National Parks, and the necessity to improve habitat protection has been recognized (Campbell et al. 2008). The global captive population of silvery gibbons totals 119 individuals, 71 of which are located in zoological institutions in Indonesia, where attempts to breed the species have been largely unsuccessful (Campbell et al. 2008; Cocks 2000). Outside of Indonesia, however, there are 48 individuals spread across several zoological institutions and 24 animals at Port Lympne and Howletts parks within the UK (Stanbury 2015). Port Lympne and Howletts zoos have selected the species for an overall conservation program that includes both a captive breeding and re-introduction program. The International Union for Conservation of Nature (IUCN) recommends that conservation should be carried out at three levels, the ecosystem, the species, and the genetic level (Frankham 2010). For the latter purpose, MHC typing appears to be appropriate, and we therefore wanted to establish the DRB-STR typing method for this endangered species on DNA isolated from feces for animal welfare reasons. So far, nothing was known about the polymorphism and variability of the DRB genes of the silvery gibbon (Hymo-DRB). Fortunately, primers that had been developed for the amplification of the human DRB-STR could be successfully used for the amplification of Hymo- as well as Paan-DRB. For the latter, primers used for the amplification of the DRB-STR in macaques could additionally be employed. In such a way, a large cohort of olive baboons, which had been partially pedigreed beforehand, has been defined for their DRB content. Although it has been widely reported that both DNA yield and quality derived from fecal samples is lower than that extracted from blood or tissue (Chaves et al. 2006; Marrero et al. 2009; Wasser et al. 1997), feces-extracted DNA of 20 of 21 silvery gibbons, housed at Port Lymphe and Howletts parks, allowed DRB-STR typing as well. DRB genotyping and haplotyping results of olive baboons and silvery gibbons will be presented. The data open opportunities for MHC analysis in captured and endangered primates in the context of animal welfare, and the impact on colony management and/or conservation biology will be discussed.
Material and methods
Animals
Olive baboon
Olive baboons, originally obtained from the Centre National de la Recherche Scientifique Centre de Primatologie (Rousset, France), were housed at the large animal facility of the INSERM unit 1064 (Nantes, France). Blood sampling was performed under anesthesia in accordance with the institutional ethical guidelines. The 154 animals analyzed in this study were partly pedigreed and descended from 34 sires and 105 dames. In most cases, the sires were known but often two males could possibly have sired the offspring.
Silvery gibbon
Twenty-four silvery gibbon individuals reside at Port Lympne and Howletts, UK, zoological institutions, of which the pedigree was partly known. Fecal samples were allocated to specific individuals either by direct observation, and two fecal samples per individual were collected immediately or by hand feeding foodstuffs containing maize that could then be identified on subsequent days. Samples were immediately frozen upon collection.
DNA extraction from EDTA blood of olive baboons
DNA was extracted from fresh EDTA or citrate blood samples using a conventional phenol/chloroform method. Briefly, white blood cells (WBC) were harvested after the lysis of red blood cells and removing the plasma by spinning down the WBCs. WBC pellets were then lysed using a 0.2% SDS and 100 μg/ml proteinase K solution and incubated overnight at 55 °C. An equal volume of phenol was then added to the lysate, and the incubation tubes were intensively mixed and then centrifuged. The upper aqueous layer was harvested and an equal volume of phenol and then of chloroform were added. Tubes were again intensively mixed, centrifuged, and the upper aqueous layer was harvested. Finally, an equal volume of chloroform was added, tubes were mixed, centrifuged, and the upper aqueous layer was harvested again. DNA were precipitated at −20 °C with two volumes of 96% ethanol. The DNA pellets were washed using 70% ethanol and then air-dried before reconstitution in sterile water. DNA concentration were adjusted at 50 ng/μl and stored at 4 °C.
DNA extraction from fecal samples of the silvery gibbon
DNA was extracted from the frozen fecal samples using the QIAamp DNA stool Mini Kit (Qiagen) following the Stool Larger Volumes protocol. This commercially available kit was chosen as it included a step that involved binding secondary compounds found in plant matter that is present in herbivorous diets, which is applicable to the silvery gibbon species. Plant secondary compounds negatively impact the PCR process by interfering with the Taq polymerase enzymatic reaction (Marrero et al. 2009). For each DNA extraction, an amount of 400 mg of frozen stool was used ensuring that both internal and external surfaces of the feces were present as a precautionary measure as sloughed epithelial cells may not be homogenously distributed throughout the sample (Piggott MaT 2003).
DRB-STR genotyping
Amplification of the relevant DNA segment in olive baboon and silvery gibbon was performed as described for humans and rhesus macaques using the same primer sets (Doxiadis et al. 2007). Briefly, the relevant DNA was amplified with a labeled forward primer located at the end of exon 2, 5′HLA-DRB-STR_VIC and a reverse primer in intron 2, namely 3′HLA-DRB-STR or the unlabeled primer 5′Mamu-DRB-STR together with the labeled macaque specific primer 3′Mamu-DRB-STR_VIC. The labeled primers were synthesized by Applied Biosystems (Foster City, USA) and the unlabeled primers by Invitrogen (Paisley, Scotland). The PCR reaction was performed in a 25-μl reaction volume containing 1 unit of Taq polymerase (Invitrogen, Paisley, Scotland) with 0.1 μM of the forward primer, 0.1 μM of the reverse primer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 1× PCR buffer II (Invitrogen, Paisley, Scotland), and 100 ng DNA.
The cycling parameters were a 5-min 94 °C initial denaturation step, followed by 5 cycles of 1 min at 94 °C, 45 s at 58 °C, and 45 s at 72 °C. Then the program was followed by 25 cycles with 45 s at 94 °C, 30 s at 58 °C, and 45 s at 72 °C. A final extension step was performed at 72 °C for 30 min. The amplified DNA was prepared for genotyping according to the manufacturer’s guidelines using the GeneScan™ 350Rox™ Size Standard and analyzed on an ABI 3130 genetic analyzer (Applied Biosystems). STR analysis was performed with the Genemapper software 5 program (Applied Biosystems) and all samples were at least analyzed twice. For Paan-DRB genotyping both primer pairs, HLA-DRB-STR and Mamu-DRB-STR were used. STRs defined by the HLA primers are named STR-H, and those detected by the macaque primers are called STR-M. Allele bins are defined beforehand.
PCR, cloning, and sequencing
Forty-four different Paan-DRB alleles and 15 different Hymo-DRB alleles were sequenced from exon 2 to intron 2, including the microsatellite. Therefore, we used the same primers and PCR reaction as described for humans and rhesus macaques (Doxiadis et al. 2007), and at least 48 clones per animal have been picked and sequenced. All the gained Paan-DRB and Hymo-DRB sequences were unreported and have been deposited in the EMBL database. In addition the sequences are officially designated by the IPD/MHC database (de Groot et al. 2012; Ellis et al. 2006; Robinson et al. 2003).
Phylogenetic analyses
Multiple sequence alignments of exon 2 of silvery gibbon DRB sequences together with some DRB sequences from humans and chimpanzees were performed using MacVector™ version 12.7.5 (Oxford Molecular Group) and phylogenetic analyses were then performed using MEGA version 4.0.2. Pairwise distances were calculated using maximum likelihood and Kimura-2 parameter for creating a phylogram. Confidence estimates of grouping were calculated according to the bootstrap method generated from 1000 replicates and the tree was rooted with Caja-DRB*W16:01.
Data accessibility
DNA sequences are deposited to the EMBL gene bank with accessions numbers JQ666205–JQ666210, JQ666212–JQ666215, JQ666217–JQ666230, JQ666232–JQ666251, KJ701253–KJ701266, and LN867601).
Results
Paan-DRB-STR typing
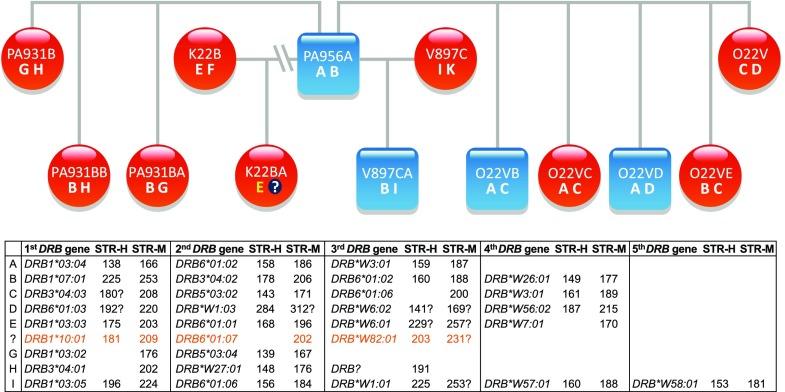
A large cohort of olive baboons (# 154) of mostly two generations originating from 34 sires and 105 dames have been analyzed with the DRB-STR method. As has been observed for other primates, microsatellite typing of the baboon samples with this STR resulted in the definition of various microsatellite patterns. As comparison, Fig. 1 shows the DRB region configurations of humans (Fig. 1a), chimpanzees (Fig. 1b), and Indian rhesus macaques (Fig. 1c), the latter of which the DRB-STR patterns are given in addition. Depending on the animal, the number of the different STR lengths in olive baboons varied from four to ten. According to co-segregation of certain microsatellite lengths within a family, patterns can be deduced which appear to represent DRB haplotypes. As has been shown in earlier studies for other primate species, each DRB allele has its own characteristic microsatellite length. Sequencing of DRB exon 2 together with the microsatellite-containing intron 2 verified that indeed each DRB allele is accompanied by a specific STR length. An example of such a DRB haplotype distribution within a baboon family is shown in the pedigree of parts of a large baboon family, and nine different haplotypes could be defined with numbers of DRB loci/genes per haplotype varying from three to five (Fig. 2, haplotypes A–I). In most cases, the DRB-STR could be defined with the macaque primers (Fig. 2, STR-M), but sometimes amplification was successful using the human primers instead (Fig. 2, STR-H). All DRB genes could be identified unambiguously by sequencing with one exception only (Fig. 2 haplotype H, STR-H 191). Likewise, there was one DRB allele, of which the STR could not be determined (Fig. 2, haplotype D, DRB*W6:02). These infrequent amplification failures of DRB alleles or STR sequences are most probably due to primer inconsistencies. Although dames and sires of the first generation were mostly not available for the analyses, it can be shown that sire PA956A has not fathered offspring K22BA, since this animal does not share one of the sire’s DRB haplotypes.
Fig. 2.
Segregation of DRB haplotypes (A–I) within an olive baboon family. The DRB alleles and their adjacent DRB-STR lengths, which define haplotypes A–I, are given below. Blue squares = males, red circles = females, question mark = DRB haplotype defined in animal K22BA, which could not be detected in sire PA956A
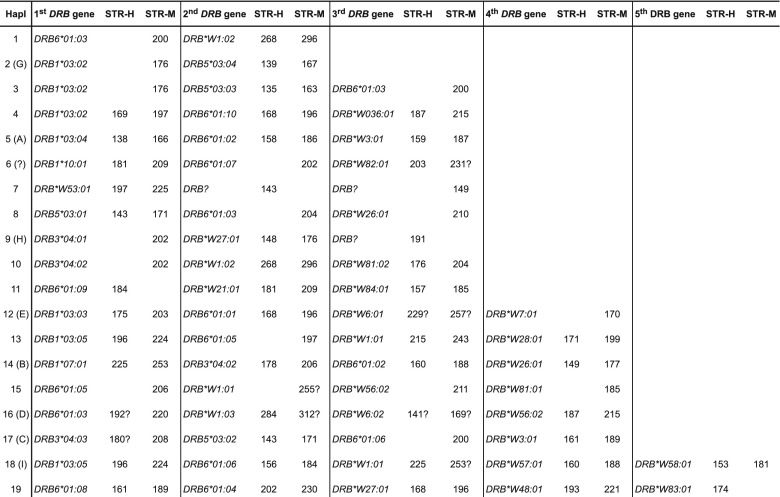
Based on the DRB typing results of the entire olive baboon panel, we have been able to define 19 different DRB region configurations/haplotypes varying in content and numbers of DRB genes present (Fig. 3). Configurations/haplotypes that have already been observed in the baboon family (Fig. 2) are indicated by the corresponding letters (e.g., Hapl. 2 in Fig. 3 is the same as Hapl. G in Fig. 2). No allelic variation within a given region configuration could be detected. Together with five DRB alleles that have been observed in a single haplotype only and are therefore not included in Fig. 2, a total of 50 Paan-DRB alleles have been newly defined (Suppl. Table 1), which belong to a high variety of lineages, and amino acid sequences have been deduced (Suppl. Table 2). Eleven of the Paan-DRB sequences are identical to a DRB allele of the Chacma baboon (Paur-DRB), the only baboon species of which many DRB alleles had been sequenced in the past (Huchard et al. 2008) (Suppl. Table 3). This sharing of DRB alleles within the genus Papio has been expected due to the trans-specific mode of evolution (Klein et al. 1993; Perelman et al. 2011). As has been observed in all apes and Old World monkey species studied so far, the number of DRB genes per haplotype may vary, in the olive baboon between two to five. Additionally to the one DRB allele of the baboon family, which could not be defined (Fig. 2, haplotype H, STR-H 191, STR-M 215), two other DRB alleles could not be determined although STRs have been identified (Fig. 3, haplotype 7). As discussed above, the rare failure to amplify certain alleles is most probably due to primer inconsistencies. As in macaques, which nearly show any allelic variation within region configurations, no allelic variation can be observed at all within a certain Paan-DRB region configuration, and thus, the number of Paan-DRB region configurations is identical to the number of different DRB haplotypes. As has been shown for other primate species, each Paan-DRB haplotype has its own characteristic DRB-STR pattern that co-segregates within a family.
Fig. 3.
Paan-DRB haplotypes defined by exon 2 sequencing and DRB-STR genotyping. Question marks indicate that STR is not or rarely detected but presence of allele is ascertained by sequencing
Hymo-DRB-STR typing
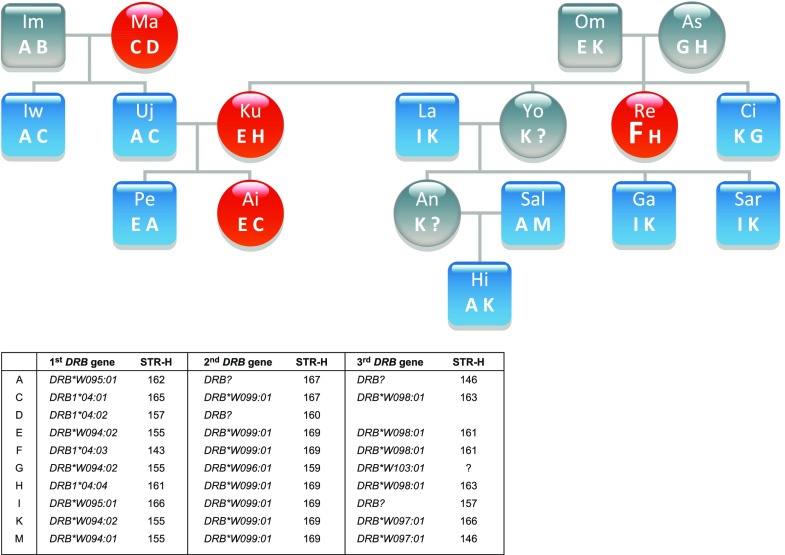
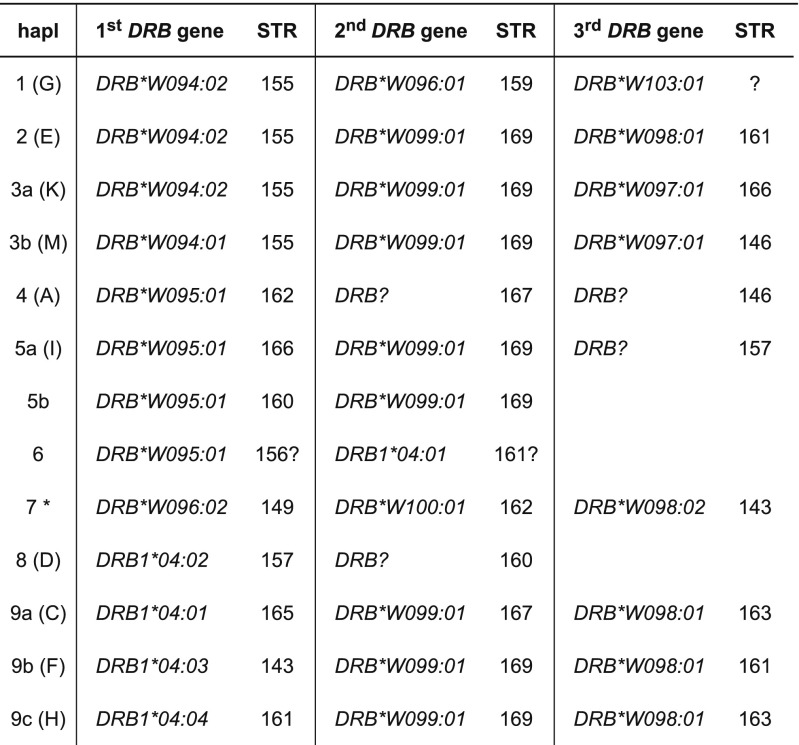
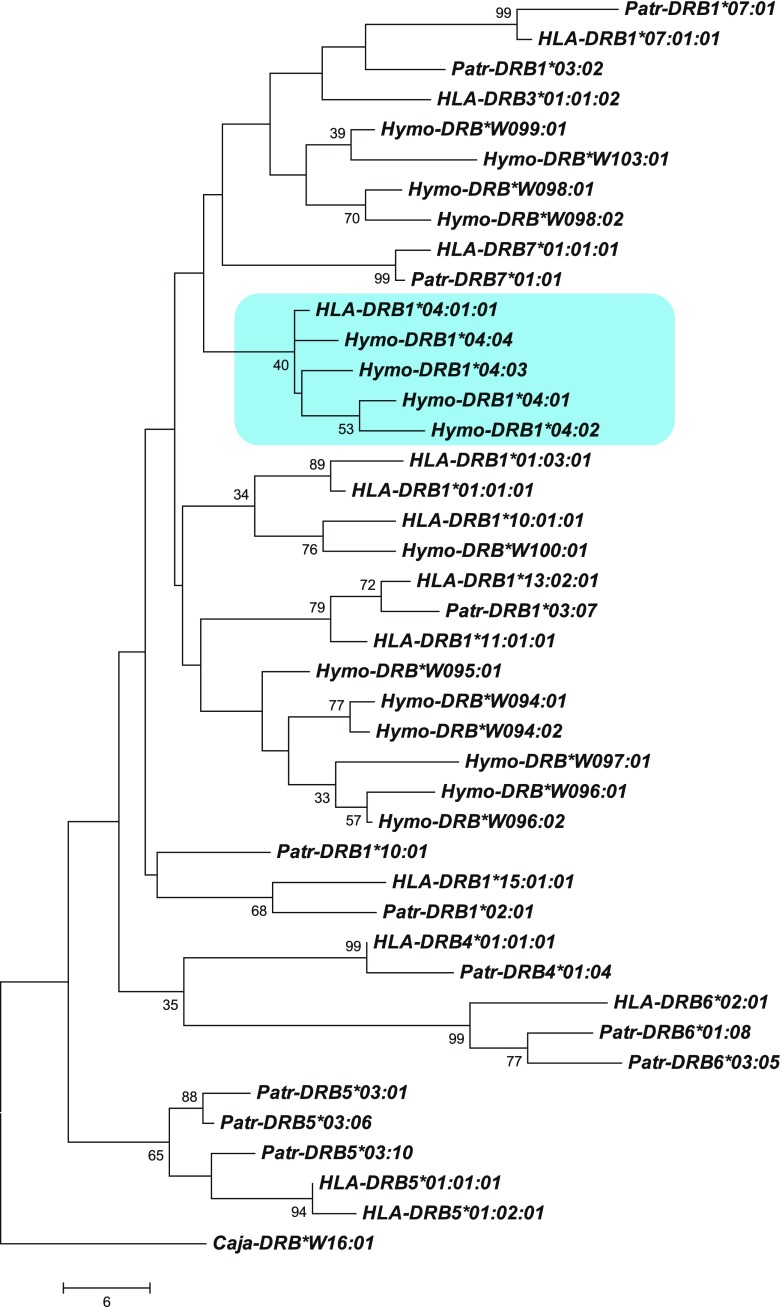
From the silvery gibbon studbook managed by Howletts zoo (Stanbury 2015) pedigree data of the families, founded by two silvery gibbon pairs and one additional female and male, were partly present for the 21 individuals of which DNA has been collected. To type for DRB of the silvery gibbon (Hymo-DRB) the same approach as for the olive baboon was followed. With the human DRB-STR primers, the DRB microsatellites could be successfully amplified of all but one silvery gibbon. As for the olive baboons, the amplification resulted in specific microsatellite patterns, which segregate in a Mendelian fashion. Subsequent sequencing of exon 2 plus the adjacent microsatellite showed that also in silvery gibbons, a certain microsatellite length is indicative for a DRB allele, and a DRB haplotype can be defined by a specific microsatellite pattern. As a result, part of the pedigree of the two founder pairs can be substantiated by the segregation of DRB haplotypes (Fig. 4, haplotypes A–M). The female child Re, however, is most probably not fathered by sire Om, since she does not share a haplotype with this male. The analysis of DRB of all animals resulted in the definition of nine region configurations (Fig. 5; region configurations, which had already been defined in the silvery gibbon’s pedigree, are indicated by letters next to haplotype numbers). Although we cannot rule out the possibility that we did not detect all DRB alleles by analyzing DNA isolated from feces, the number of DRB alleles (N = 15) and the composition of the DRB region configurations/haplotypes appears to show a low degree of variability. In contrast to other primate species and especially to Old World monkeys as baboons (Fig. 3), most Hymo-DRB haplotypes seem to be constructed by three DRB loci. In addition, the DRB alleles belong to only a small number of DRB lineages but show instead allelic variation within a certain region configuration. As a consequence, 13 different DRB haplotypes could be determined, most of which consist of combinations of alleles of the same, few lineages (Fig. 5, e.g., DRB1*04; DRB*W094, DRB*W098). Furthermore, the lineage DRB*W099 seems to be monomorphic and its one allele, DRB*W099:01, is present on 8 of 13 haplotypes. Remarkably, the DRB5 and DRB6 lineages, which are both old entities in primate evolution, are not observed (Doxiadis et al. 2012). However, we cannot exclude that those alleles, which could not be amplified with the primers used (Fig. 5, “?”), probably belong to these lineages. Phylogenetic analysis of exon 2 of Hymo-DRB alleles together with a selection of human and chimpanzee DRB alleles shows that the silvery gibbon alleles defined in this study cluster into two species-specific branches with two exceptions; first, Hymo-DRB*W100:01 groups apart together with a human DRB1*10 lineage member; the second exception concerns the Hymo-DRB1*04 alleles which cluster together with HLA-DRB1*04:01:01 (Fig. 6). From previous studies, we know that the HLA-DRB1*04 lineage is absent in the chimpanzee and probably also in other ape species and represents together with HLA/Patr-DRB1*07 and HLA-DRB1*09 an evolutionarily old entity (Doxiadis et al. 2012). It is therefore remarkable that the DRB1*04 lineage is retained in silvery gibbons, an observation, which is confirmed by an identical motif within the peptide binding groove (Suppl. Fig. 1, amino acids EQVKH, turquois) in silvery gibbons, humans, and even in rhesus macaques. The intron 2 microsatellite sequences of Hymo-DRB1*04 alleles, however, are different from the human DRB1*04 sequence (Suppl. Fig. 2) indicating that intron sequences may vary between species.
Fig. 4.
Segregation of DRB haplotypes (A–M) within a silvery gibbon family. The DRB alleles and their adjacent DRB-STR lengths, which define haplotypes A–M, are given below. Blue squares = males, red circles = females, question mark = DRB haplotype that could not been defined, F = haplotype of female Re which is not present in sire Om
Fig. 5.
Hymo-DRB haplotypes defined by exon 2 sequencing and DRB-STR genotyping. Asterisk = haplotype not certain. DRB-STR 161? is detected but could not be confirmed
Fig. 6.
Phylogenetic tree of DRB exon 2 sequences of humans (HLA-DRB), chimpanzees (Patr-DRB), and silvery gibbons (Hymo-DRB)
Discussion
In two non-human primate species, for which no MHC class II data have been available so far, the analysis of the Mhc-DRB allele content has been effectively performed by the presented robust, time-saving, and inexpensive microsatellite typing technique. In such a way, the DRB repertoire of an olive baboon population could be defined. The results obtained are of importance for biomedical research as in transplantation (Le Bas-Bernardet et al. 2015; Poirier et al. 2015), for which these animals are a model species. Due to the high CNV of the DRB genes in baboons and the possibility to define DRB haplotypes, the DRB content in these animals can be followed in addition for colony management reasons as parentage testing and calculation of the inbreeding coefficient.
As has been shown previously for a wild Assamese macaque population (Muller et al. 2014), DRB-STR typing was also successful when DNA isolated from non-invasively collected fecal material was used. This is especially important in the case of wild or free ranging endangered species such as the silvery gibbon. The quality of the isolated DNA was high, most probably due to the immediate freezing of the sample. As a consequence, cloning and sequencing of DRB exon 2 including the microsatellite was feasible, and in such a way the DRB-STR length could be linked to a certain DRB allele. Therefore, we were able to get an insight into the DRB repertoire of the silvery gibbon, which shows an important reduction in the number of lineages present and in CNV in comparison to chimpanzees, humans, and especially to the olive baboon analyzed. Due to the existence of several, mostly three, DRB genes with adjacent microsatellites per haplotype, DRB haplotypes could be defined for the silvery gibbon, too. Although the genetic repertoire as defined for Hymo-DRB appears to be reduced, we were able to confirm the pedigree of two related founder families as recorded in the studbook. In addition, results with this one microsatellite indicated that the father of one of the animals was incorrectly assigned.
Despite their conservation importance, distinct characteristics and high species diversity gibbons are mostly neglected by population genetic studies (Kim et al. 2011). However, several reports describe the high rates of chromosomal rearrangements that can be observed not only in the large number of rearrangements separating gibbons from other apes and humans (Carbone et al. 2006; Muller et al. 2014) but also the numerous rearrangements that separate different gibbon species from each other as well as rearrangements that are polymorphic within a species (Carbone et al. 2009; Van Tuinen et al. 1999). The possible mechanism for this genomic plasticity may be a gibbon-specific retrotransposon, LAVA (Carbone et al. 2014). The observed “patchwork” pattern of most of the DRB haplotypes of the silvery gibbon, which are built up of three DRB genes of only a few lineages, may also be the result of a near-instantaneous radiation (Carbone et al. 2014) and rearrangements in the DRB region of this species.
The complex microsatellite in DRB intron 2 is an evolutionarily old entity, which has been observed not only in apes and OWM but also in Platyrrhini species as in marmosets and owl monkeys (Trtkova et al. 1995), and it is observed even in sheep (Ballingall et al. 2008). In the two Aotus species it is highly polymorphic and has been used for DRB typing, too (Lopez et al. 2014). Thus, this typing method is applicable for nearly all primates. Since DRB-STR typing is highly informative and can additionally be easily performed by feces collection, this methodology can be used to observe differences in genetic fitness between wild and free-ranging animals. Additionally, this method is applicable for monitoring, e.g., reproduction, parentage typing, mate choice, parasite susceptibility, and conservation. Furthermore, in the case of breeding of non-human primates, the DRB microsatellite is valuable for parentage typing, avoidance of inbreeding and genetic reduction, and thus for colony management in general.
Electronic supplementary material
(PDF 71 kb)
(PDF 32 kb)
(PDF 32 kb)
Alignment of deduced amino acids of part of DRB exon 2 of humans, chimpanzees, rhesus macaques, and silvery gibbons. The alignment is sorted according the amino acids (9–13) of the peptide binding site. (PDF 27 kb)
Nucleotide composition of the DRB-STR in intron 2 of humans, chimpanzees, and silvery gibbons (PDF 54 kb)
Acknowledgements
The authors wish to thank Francisca van Hassel for preparing the figures. The work was supported in part by NIH/NIAID project HHSN272201100013C.
Compliance with ethical standards
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals are followed.
References
- Andersson G, Larhammar D, Widmark E, Servenius B, Peterson PA, Rask L. Class II genes of the human major histocompatibility complex. Organization and evolutionary relationship of the DR beta genes. J Biol Chem. 1987;262:8748–8758. [PubMed] [Google Scholar]
- Ballingall KT, Fardoe K, McKeever DJ. Genomic organisation and allelic diversity within coding and non-coding regions of the Ovar-DRB1 locus. Immunogenetics. 2008;60:95–103. doi: 10.1007/s00251-008-0278-2. [DOI] [PubMed] [Google Scholar]
- Bergstrom TF, Erlandsson R, Engkvist H, Josefsson A, Erlich HA, Gyllensten U. Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol Rev. 1999;167:351–365. doi: 10.1111/j.1600-065X.1999.tb01404.x. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Cai R, Shafer AB, Laguardia A, Lin Z, Liu S, Hu D. Recombination and selection in the major histocompatibility complex of the endangered forest musk deer (Moschus berezovskii) Sci Rep. 2015;5:17285. doi: 10.1038/srep17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C, Andayani N, Cheyne S, Pamungkas J, Manullang B, Usman F, Wedana M, Traylor-Holzer K (eds) (2008) Indonesian gibbon conservation and management workshop final report. IUCN/SSC conservation breeding specialist group, Apple Valley, MN
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B, Anaclerio F, Archidiacono N, Baker C, Barrell D, Batzer MA, Beal K, Blancher A, Bohrson CL, Brameier M, Campbell MS, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513:195–201. doi: 10.1038/nature13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J, Milosavljevic A, de Jong PJ. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:e1000538. doi: 10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BF, Zhu B, Osoegawa K, Mootnick A, Kofler A, Wienberg J, Rogers J, Humphray S, Scott C, Harris RA, Milosavljevic A, de Jong PJ. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006;2:e223. doi: 10.1371/journal.pgen.0020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves P, Paes MF, Mendes SL, Strier KB, Louro ID, Fagundes V. Noninvasive genetic sampling of endangered muriqui (primates, Atelidae): efficiency of fecal DNA extraction. Genet Mol Biol. 2006;29:750–754. doi: 10.1590/S1415-47572006000400028. [DOI] [Google Scholar]
- Cocks LR. International studbook for the silvery gibbon (Hylobates moloch) Western Australia: Perth Zoo; 2000. [Google Scholar]
- de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE. Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics. 2008;60:737–748. doi: 10.1007/s00251-008-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh SG, O’Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE. Nomenclature report on the major histocompatibility complex genes and alleles of great ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, Claas FH, Doxiadis II, van Rood JJ, Bontrop RE. A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc Natl Acad Sci U S A. 2007;104:8907–8912. doi: 10.1073/pnas.0702964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, Otting N, de Vos-Rouweler AJ, Bolijn MJ, Heijmans CM, de Groot NG, van der Wiel MK, Remarque EJ, Vangenot C, Nunes JM, Sanchez-Mazas A, Bontrop RE. Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics. 2013;65:569–584. doi: 10.1007/s00251-013-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, Hoof I, de Groot N, Bontrop RE. Evolution of HLA-DRB genes. Mol Biol Evol. 2012;29:3843–3853. doi: 10.1093/molbev/mss186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, Stet RJ, Waller MJ, Walter L, Marsh SG. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- Epplen C, Santos EJ, Guerreiro JF, van Helden P, Epplen JT. Coding versus intron variability: extremely polymorphic HLA-DRB1 exons are flanked by specific composite microsatellites, even in distant populations. Hum Genet. 1997;99:399–406. doi: 10.1007/s004390050379. [DOI] [PubMed] [Google Scholar]
- Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv. 2010;143:1919–1927. doi: 10.1016/j.biocon.2010.05.011. [DOI] [Google Scholar]
- Hans JB, Haubner A, Arandjelovic M, Bergl RA, Funfstuck T, Gray M, Morgan DB, Robbins MM, Sanz C, Vigilant L. Characterization of MHC class II B polymorphism in multiple populations of wild gorillas using non-invasive samples and next-generation sequencing. Am J Primatol. 2015;77:1193–1206. doi: 10.1002/ajp.22458. [DOI] [PubMed] [Google Scholar]
- Huchard E, Weill M, Cowlishaw G, Raymond M, Knapp LA. Polymorphism, haplotype composition, and selection in the Mhc-DRB of wild baboons. Immunogenetics. 2008;60:585–598. doi: 10.1007/s00251-008-0319-x. [DOI] [PubMed] [Google Scholar]
- Kim SK, Carbone L, Becquet C, Mootnick AR, Li DJ, de Jong PJ, Wall JD. Patterns of genetic variation within and between gibbon species. Mol Biol Evol. 2011;28:2211–2218. doi: 10.1093/molbev/msr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Satta Y, O’HUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Le Bas-Bernardet S, Tillou X, Branchereau J, Dilek N, Poirier N, Chatelais M, Charreau B, Minault D, Hervouet J, Renaudin K, Crossan C, Scobie L, Takeuchi Y, Diswall M, Breimer ME, Klar N, Daha MR, Simioni P, Robson SC, Nottle MB, et al. Bortezomib, C1-inhibitor and plasma exchange do not prolong the survival of multi-transgenic GalT-KO pig kidney xenografts in baboons. Am J Transplant. 2015;15:358–370. doi: 10.1111/ajt.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C, Suarez CF, Cadavid LF, Patarroyo ME, Patarroyo MA. Characterising a microsatellite for DRB typing in Aotus vociferans and Aotus nancymaae (Platyrrhini) PLoS One. 2014;9:e96973. doi: 10.1371/journal.pone.0096973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero P, Fregel R, Cabrera VM, Nogales M. Extraction of high-quality host DNA from feces and regurgitated seeds: a useful tool for vertebrate ecological studies. Biol Res. 2009;42:147–151. doi: 10.4067/S0716-97602009000200002. [DOI] [PubMed] [Google Scholar]
- Muller N, Ostner J, Schulke O, Walter L. Towards the non-invasive assessment of MHC genotype in wild primates: analysis of wild Assamese macaque MHC-DRB from fecal samples. Am J Primatol. 2014;76:230–238. doi: 10.1002/ajp.22225. [DOI] [PubMed] [Google Scholar]
- Nijman V. In-situ and ex-situ status of the Javan gibbon and the role of zoos in conservation of this species. Contrib Zool. 2004;75:161–168. [Google Scholar]
- Nishita Y, Abramov AV, Kosintsev PA, Lin LK, Watanabe S, Yamazaki K, Kaneko Y, Masuda R. Genetic variation of the MHC class II DRB genes in the Japanese weasel, Mustela itatsi, endemic to Japan, compared with the Siberian weasel, Mustela sibirica. Tissue Antigens. 2015;86:431–442. doi: 10.1111/tan.12700. [DOI] [PubMed] [Google Scholar]
- Oliver MK, Piertney SB. Selection maintains MHC diversity through a natural population bottleneck. Mol Biol Evol. 2012;29:1713–1720. doi: 10.1093/molbev/mss063. [DOI] [PubMed] [Google Scholar]
- Oliver MK, Telfer S, Piertney SB. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris) Proc Biol Sci. 2009;276:1119–1128. doi: 10.1098/rspb.2008.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AJ, Pearson J, Negro SS, Chilvers BL, Kennedy MA, Gemmell NJ. Heterozygote advantage at MHC DRB may influence response to infectious disease epizootics. Mol Ecol. 2015;24:1419–1432. doi: 10.1111/mec.13128. [DOI] [PubMed] [Google Scholar]
- Pechouskova E, Dammhahn M, Brameier M, Fichtel C, Kappeler PM, Huchard E. MHC class II variation in a rare and ecological specialist mouse lemur reveals lower allelic richness and contrasting selection patterns compared to a generalist and widespread sympatric congener. Immunogenetics. 2015;67:229–245. doi: 10.1007/s00251-015-0827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O’Brien SJ, Pecon-Slattery J. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- Piggott MaT AC. Extensive evaluation of faecal preservation and DNA extraction methods in Australian native and introduced species. Australian Journal of Zoology. 2003;51:341–355. doi: 10.1071/ZO03012. [DOI] [Google Scholar]
- Poirier N, Dilek N, Mary C, Ville S, Coulon F, Branchereau J, Tillou X, Charpy V, Pengam S, Nerriere-Daguin V, Hervouet J, Minault D, Le Bas-Bernardet S, Renaudin K, Vanhove B, Blancho G. FR104, an antagonist anti-CD28 monovalent fab′ antibody, prevents alloimmunization and allows calcineurin inhibitor minimization in nonhuman primate renal allograft. Am J Transplant. 2015;15:88–100. doi: 10.1111/ajt.12964. [DOI] [PubMed] [Google Scholar]
- Poirier N, Mary C, Le Bas-Bernardet S, Daguin V, Belarif L, Chevalier M, Hervouet J, Minault D, Ville S, Charpy V, Blancho G, Vanhove B. Advantages of Papio anubis for preclinical testing of immunotoxicity of candidate therapeutic antagonist antibodies targeting CD28. MAbs. 2014;6:697–707. doi: 10.4161/mabs.28375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess O, Kammerbauer C, Roewer L, Steimle V, Andreas A, Albert E, Nagai T, Epplen JT. Hypervariability of intronic simple (gt)n(ga)m repeats in HLA-DRB genes. Immunogenetics. 1990;32:110–116. doi: 10.1007/BF00210448. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setchell JM, Abbott KM, Gonzalez JP, Knapp LA. Testing for post-copulatory selection for major histocompatibility complex genotype in a semi-free-ranging primate population. Am J Primatol. 2013;75:1021–1031. doi: 10.1002/ajp.22166. [DOI] [PubMed] [Google Scholar]
- Setchell JM, Vaglio S, Abbott KM, Moggi-Cecchi J, Boscaro F, Pieraccini G, Knapp LA. Odour signals major histocompatibility complex genotype in an Old World monkey. Proc Biol Sci. 2011;278:274–280. doi: 10.1098/rspb.2010.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbury K. Supplementing the studbook. Using genetic analysis to complement a captive breeding programme of an endangered primate, Hylobates moloch. United Kingdom: Writtle College, University of Essex; 2015. [Google Scholar]
- Trtkova K, Mayer WE, O’Huigin C, Klein J. Mhc-DRB genes and the origin of New World monkeys. Mol Phylogenet Evol. 1995;4:408–419. doi: 10.1006/mpev.1995.1038. [DOI] [PubMed] [Google Scholar]
- Van Tuinen P, Mootnick AR, Kingswood SC, Hale DW, Kumamoto AT. Complex, compound inversion/translocation polymorphism in an ape: presumptive intermediate stage in the karyotypic evolution of the agile gibbon Hylobates agilis. Am J Phys Anthropol. 1999;110:129–142. doi: 10.1002/(SICI)1096-8644(199910)110:2<129::AID-AJPA2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wasser SK, Houston CS, Koehler GM, Cadd GG, Fain SR. Techniques for application of faecal DNA methods to field studies of ursids. Mol Ecol. 1997;6:1091–1097. doi: 10.1046/j.1365-294X.1997.00281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 71 kb)
(PDF 32 kb)
(PDF 32 kb)
Alignment of deduced amino acids of part of DRB exon 2 of humans, chimpanzees, rhesus macaques, and silvery gibbons. The alignment is sorted according the amino acids (9–13) of the peptide binding site. (PDF 27 kb)
Nucleotide composition of the DRB-STR in intron 2 of humans, chimpanzees, and silvery gibbons (PDF 54 kb)
Data Availability Statement
DNA sequences are deposited to the EMBL gene bank with accessions numbers JQ666205–JQ666210, JQ666212–JQ666215, JQ666217–JQ666230, JQ666232–JQ666251, KJ701253–KJ701266, and LN867601).